Federal Court of Australia
ToolGen Incorporated v Fisher (No 2) [2023] FCA 794
Table of Corrections | |
[95] and [96] delete the word “Type II” where appearing in those paragraphs | |
[196] replace reference to (f)-(g) with (a)-(b) | |
[260] and [261] corrections to formatting | |
[334] replace reference to (d)-(j) with (a)-(g) | |
[408] insert the words “in relation to P1” in penultimate sentence and the words “they say” in the last sentence |
ORDERS
Appellant | ||
AND: | First Respondent ACN 004 552 363 PTY LTD Second Respondent | |
AND BETWEEN: | First Cross-Appellant ACN 004 552 363 PTY LTD Second Cross-Appellant | |
AND: | Cross-Respondent | |
DATE OF ORDER: |
THE COURT ORDERS THAT:
1. The appellant file and serve any interlocutory application seeking an order directing that the complete specification be amended pursuant to s 105(1A) of the Patents Act 1990 (Cth) together with any affidavit in support of such application by 4.00pm, 11 August 2023.
2. The proceeding be stood over to 9.30am on 17 August 2023 for the making of:
(a) final orders in the event no application is filed pursuant to order 1;
(b) for the making of further orders in relation to any application filed pursuant to order 1; and
(c) other orders (including in relation to costs) as may be considered appropriate.
Note: Entry of orders is dealt with in Rule 39.32 of the Federal Court Rules 2011.
REASONS FOR JUDGMENT
[1] | |
[10] | |
[14] | |
[20] | |
[20] | |
[21] | |
[25] | |
[30] | |
[33] | |
[35] | |
[36] | |
[46] | |
[47] | |
[49] | |
[51] | |
[52] | |
[56] | |
[56] | |
[73] | |
[76] | |
[76] | |
[82] | |
[91] | |
[97] | |
[101] | |
[113] | |
[113] | |
[146] | |
[160] | |
[167] | |
[194] | |
[199] | |
A Type II CRSIPR/Cas system from a bacterial species other than S. pyogenes | [213] |
[366] | |
[371] | |
[377] | |
[379] | |
[383] | |
[391] | |
[412] | |
[415] | |
[423] | |
[432] | |
[434] |
1 This proceeding concerns an opposed patent application for what is now a well-known gene editing system known as the CRISPR/Cas9 system. The CRISPR/Cas9 system described in the application can be used to edit target DNA sequences in eukaryotic cells so as to disable or modify gene expression through (inter alia) the deletion or insertion of such sequences using an RNA-guided endonuclease.
2 The appellant (“ToolGen”) is the applicant in Australian Patent Application 2013335451 (“the patent application”). The patent application relates to compositions and methods involving a system for introducing a site-specific double-stranded break (or cleavage) at a target nucleic acid sequence in a eukaryotic cell comprising a nucleic acid encoding a Cas9 polypeptide and a nucleic acid encoding a guide RNA specific for the target DNA. The system disclosed in the patent application is referred to as a “Type II Clustered Regularly Interspaced Short Palindromic Repeats/Cas system” or “CRISPR/Cas system”. The patent application has 21 claims including independent claim 1 for a composition and independent claim 10 for a method.
3 The patent application was filed on 23 October 2013 and relies on an earliest priority date of 23 October 2012 based on US Provisional Patent Application 61/717,324 (“P1”). There are two other priority documents referred to in the patent application being US Provisional Patent Application 61/803/599 (“P2”) with a filing date of 20 March 2013 and US Provisional Patent Application 61/837,481 (“P3”) with a filing date of 20 June 2013. No submissions were made by either party in relation to P2 or P3 and none of the experts were questioned about them. I will say a little more about them later in these reasons. It is sufficient to say at this point that P2 is incapable of conferring priority on any claim in the patent application and P3 was filed after the publication date of various journal articles that deprive the claims of novelty or any inventive step.
4 The first respondent opposed the patent application before the Commissioner of Patents. That opposition was successful in relation to claims 1-8 and 10-18 of the patent application, which the Delegate of the Commissioner of Patents (“Delegate”) found were not novel and did not involve an inventive step in circumstances where none of those claims was entitled to priority from P1. Claim 19 was found to lack clarity. Claim 21 was also found to not involve an inventive step. The Delegate indicated that she would allow ToolGen two months to propose appropriate amendments. (See Fisher v ToolGen Inc (2018) 144 IPR 315, [2018] APO 65).
5 ToolGen appealed the Delegate’s decision pursuant to s 60(4) of the Patents Act 1990 (Cth) (“the Act”). The second respondent was named as an additional respondent. The respondents filed a cross-appeal in relation to claims 9 and 20. They have also raised additional grounds of invalidity which were rejected by the Delegate.
6 Even though this proceeding is referred to as an appeal, it is well-established that it is not an appeal in the strict sense but is conducted as a hearing de novo in the original jurisdiction of the Court: Commissioner of Patents v Sherman (2008) 172 FCR 394 at [18].
7 It is common ground that if claims 1-8 and 10-18 are not entitled to priority from P1, they are not novel and lack an inventive step. As to claims 9 and 19-21, the respondents contend that they also lack novelty and do not involve an inventive step if they are not entitled to priority from P1.
8 In these reasons I refer to various publications in the scientific literature. Sometimes I refer to these publications by their full citation but more often than not it is sufficient to identify them by the lead authors name and the year of publication (eg. Wang (2013)). Full details of these publications are set out in the Bibliography in Annexure A to these reasons.
9 The Primer (Exhibit A) which was agreed between the parties was of considerable assistance to me. It covers a range of topics including the basics of cell biology, the genetic code, molecular biology, and gene editing. The matters described in paras 14-101 of the Primer (which I need not reproduce) are elementary and were very well known to molecular biologists before the priority date.
10 The parties agreed on a lengthy and detailed statement of issues which I found helpful and have had regard to, even though I have chosen not to frame my reasons for judgment around it.
11 Broadly stated, the principal issues addressed in these reasons are as follows:
(a) who is the skilled addressee of P1 and the patent application and what was the common general knowledge of the skilled addressee (or skilled team) as at 23 October 2012?
(b) what construction should be given to claims 1 and 10 (and their dependent claims) of the patent application, including to the phrase “nucleic acid encoding a guide RNA”?
(c) do claims 1 and 10 (and their dependent claims) extend to “paired Cas nickases”?
(d) does claim 19 of the patent application lack clarity?
(e) does P1 provide an enabling disclosure of the invention claimed in each of the claims of the patent application?
(f) if the priority date is deferred, does the invention claimed in each of claims 9, 19 and 20 of the patent application lack novelty in light of Wang (2013)?
(g) if the priority date is deferred, does the invention claimed in each of claims 9, 19, 20 and 21 of the patent application not involve an inventive step in light of the common general knowledge at the relevant date considered together with each of Cong (2013), Mali (2013) and Wang (2013) (taken separately)?
(h) does the complete specification of the patent application comply with s 40(2)(a) of the Act in respect of the invention claimed in each claim?
(i) is each claim of the patent application supported in accordance with s 40(3) of the Act by matter disclosed in the complete specification?
ToolGen accepts that if the priority date is deferred, then claims 1-8 and 10-18 will lack novelty and an inventive step. ToolGen makes no such concession in relation to claims 9, 19, 20 or 21.
12 With regard to issues (e), (h) and (i), the respondents contend that these questions should be answered in the negative because neither P1 nor the patent application discloses the invention as claimed or, alternatively, does not enable its use across the full scope of each claim. These issues raise questions as to the proper construction and application of s 40(2)(a), s 40(3) and s 43(2A) in the form they have taken since the Act was amended by the RTB Act.
13 For the reasons that follow I have concluded:
(a) None of the claims are entitled to priority based on P1 (s 43(2A)).
(b) All of the claims lack novelty or do not involve an inventive step (s 18(1)(b)).
(c) The complete specification does not provide an enabling disclosure of the invention (s 40(2)(a)).
(d) The claims are not supported by matter disclosed in the specification (s 40(3)).
(e) Claim 19 lacks clarity (s 40(3)).
14 Each party made submissions concerning the onus of proof. ToolGen emphasised that the legal burden on all issues is on the opponent. The respondents did not dispute that they carry the legal burden in this proceeding. However, they drew attention to the following observations in the Explanatory Memorandum to the Intellectual Property Laws Amendment (Raising the Bar) Act 2012 (Cth) (“RTB Act”) concerning the amendment to s 40(2)(a) of the Act and the requirement that there be an enabling disclosure:
A specification that provides a single example of the invention may satisfy the requirements, but only where the skilled person can extend the teaching of the specification to produce the invention across the full width of the claims, without undue burden, or the need for further invention.
However, it is expected to be more likely that, where the claims are broad, the specification will need to give a number of examples or describe alternative embodiments or variations extending over the full scope of the claims. This ensures that the monopoly extends only to that which could reasonably be said to be disclosed and no further.
If, on its face, the specification would appear to the skilled person to lack sufficient disclosure, the onus of establishing that the invention is described in enough detail lies with the applicant (see item 14).
15 The statement concerning onus in the Explanatory Memorandum appears to be directed to the onus at the examination stage. I note that the reference to item 14 is to a proposed amendment to s 49 of the Act. That section is concerned with acceptance of a patent request rather than the hearing and determination of any opposition to the grant of a patent following acceptance. With regard to the opposition, s 60(3A) of the Act provides:
(3A) If the Commissioner is satisfied, on the balance of probabilities, that a ground of opposition to the grant of the standard patent exists, the Commissioner may refuse the application.
16 In their submissions the respondents referred to the shifting of the evidentiary onus to ToolGen in circumstances where, in their submission, P1 does not on its face, purport to provide an enabling disclosure extending to, for example, use of a Type II CRISPR/Cas9 system derived from bacterial species other than S. pyogenes. They submitted, in effect, that in these circumstances the evidentiary onus shifted to ToolGen to establish that there was an enabling disclosure.
17 I do not consider it helpful to speak of a shifting evidential onus in this case. Ultimately, it is for the respondents to persuade the Court that P1 does not provide an enabling disclosure. To the extent it is necessary to resolve a disputed issue of fact in determining whether that objection is established, the issue is to be determined on the balance of probabilities.
18 In deciding whether there is an enabling disclosure, the Court will necessarily have regard to the content of P1 when read in light of the common general knowledge, the cogency of the evidence relied on by each side as to adequacy of the information made available, the difficulties that would be faced by the skilled addressee in seeking to perform the invention, and whether the work involved amounts to an undue burden. The determination of that question involves an evaluative judgment based on a consideration of both the nature of the technology and the work required of the skilled addressee to perform the invention across the scope of the claims.
19 Even though the burden of proof is on the respondents, circumstances may still arise in which ToolGen’s failure to adduce any evidence or any sufficient evidence on some particular matter (eg. a fact which it asserts was common general knowledge) may ultimately lead the Court to conclude, on the totality of the relevant evidence, that the invention cannot be performed across the full scope of the claims without undue burden. This may be particularly true in relation to matters in respect of which P1 is wholly silent.
Eukaryotic and prokaryotic cells
20 Eukaryotes are organisms comprised of one or more eukaryotic cells. A eukaryotic cell has a defined nucleus which is an organelle that contains DNA enclosed within a nuclear envelope (double membrane). Mammals (including humans) are classified as eukaryotes as they are comprised of eukaryotic cells. Prokaryotes are unicellular organisms comprised of a prokaryotic cell. A prokaryotic cell has no nucleus or membrane bound organelles, and DNA in prokaryotic cells is found in the form of supercoiled circular DNA that is not enclosed by a nuclear membrane. Bacteria are an example of a prokaryote.
21 DNA (deoxyribonucleic acid) and RNA (ribonucleic acid) are both nucleic acids consisting of a series (or string) of nucleotides. The nucleotides in DNA and RNA each contain a sugar, a nitrogenous base and a phosphate group. In DNA, the sugar is deoxyribose whereas in RNA the sugar is ribose. The nitrogenous base in each nucleotide in a DNA molecule is either a purine (being adenine (A) or guanine (G)) or a pyrimidine (being cytosine (C) or thymine (T)). The nitrogenous bases in RNA are the same as those in DNA except that uracil (U) is substituted for thymine (T). Nucleotides are joined together to form a long chain of DNA or RNA. This long chain is termed a polynucleotide.
22 In DNA, the sequence in which the four bases (A, C, G, T) are arranged in the polynucleotide chain comprises the DNA code. The bases in one polynucleotide chain of DNA pair with complementary bases in the other polynucleotide chain of DNA (so called “base pairing”) to form a double-stranded helical structure. In the double-stranded DNA structure, the nitrogenous base guanine (G) base pairs with cytosine (C), while the nitrogenous base adenosine (A) base pairs with thymine (T).
23 In biological cells, molecules of RNA predominantly consist of a single polynucleotide chain or strand. However, the sugar-phosphate backbone of the chain is flexible and can fold so that self-complementary sequences within the RNA pair with each other (to form a duplex, or double-stranded structure).
24 The nucleotide sequence of both DNA and RNA can be identified by a technique known as sequencing.
25 Proteins are produced (or “expressed”) in a cell by the processes of transcription (DNA to RNA) and translation (RNA to protein). In a eukaryotic cell, transcription occurs in the nucleus (where DNA is located) and translation occurs in the cytoplasm (where ribosomes are located). In a prokaryotic cell, transcription and translation occurs in the cytoplasm (where DNA and ribosomes are located). The cytoplasm is the gelatinous liquid that fills the inside of a cell that is comprised of water, salts and other organic molecules, and a ribosome is an organelle within the cytoplasm that is the site of protein synthesis.
26 During transcription, the DNA double-stranded helix unwinds and one of the two strands (the template or non-coding strand) acts as a template for the synthesis of a single-stranded RNA molecule. Transcription generates a synthesised RNA (pre-mRNA) molecule with bases complementary to the template DNA strand and with bases identical (with the exception that “U” is substituted for “T”) to the coding DNA strand. Pre-mRNA is then processed to form mature messenger RNA (mRNA).
27 During translation, mRNA acts as a template for the synthesis of a polypeptide chain (a sequence of amino acids joined together by peptide bonds) which make up a protein. The mRNA sequence is read consecutively in groups of three nucleotides, known as codons. Each codon specifies either one amino acid or comprises a start or stop codon that starts and ends the translation process.
28 There are only 20 amino acids that are commonly found in proteins, however, there are 64 possible combinations of nucleotide triplets to make up a codon (given that there are four different nucleotides which could be in each position in the triplet). This is because the same amino acid can be coded for by more than one codon. This is referred to as the degeneracy or redundancy of the genetic code.
29 The codons in a molecule of mRNA are recognised by small RNA molecules known as transfer RNA (tRNA) that are located in ribosomes. One region of the tRNA (the anticodon) binds complementarily to an mRNA codon while another region of the tRNA binds to the amino acid that matches the mRNA codon attached to the tRNA. As the mRNA sequence is read from start codon to stop codon, the amino acids coded for by the intervening codons are brought together to form a polypeptide chain (protein).
Cellular expression of proteins
30 Proteins can be expressed in eukaryotic cells (including mammalian cells) via recombinant DNA technology using vectors. Vectors can be plasmids (small circular pieces of DNA from bacteria) or phages (viruses) that transfer foreign DNA into a cell. The insertion of a foreign DNA sequence into the vector enables the DNA sequence to be propagated (cloning vectors) or used to express a protein or RNA (expression vectors).
31 The foreign DNA inserted into a vector can comprise a DNA fragment of a particular size, including DNA synthesised outside the cell (in vitro), a section of DNA from another clone to be subcloned (that is, taking a smaller part of the larger fragment), a section of DNA produced using restriction enzymes (enzymes that cut DNA) or a PCR fragment.
32 In a plasmid vector, the plasmid and foreign DNA insert are both cut with restriction enzymes which generate compatible 5’ and 3’ ends. The plasmid and insert are then combined and ligated (stitched together). The newly formed plasmid (with DNA insert) is transformed (delivered) into bacteria and selected for using antibiotic-containing growth medium.
33 As set out above, proteins are produced by the process of translation which occurs in the ribosome in the cytoplasm of cells. In eukaryotic cells, the protein must pass into the nucleus through the nuclear membrane in order for a protein to access and interact with chromosomal DNA. Nuclear proteins (ie. proteins that function in the nucleus) commonly enter the nucleus by passing through a nuclear pore channel. This can be contrasted to prokaryotic cells, where chromosomal DNA is found in the cytoplasm.
34 As of October 2012, scientists were using a number of different nuclear localisation sequences (“NLS”) as modular tags to deliver proteins or protein fragments from the cytoplasm to the nucleus. A NLS is a short peptide derived from proteins which enter the nucleus of a cell (nuclear proteins). For example, the sequence PKKKRKV is an NLS which was widely used and studied prior to October 2012.
RNA interference, Zinc finger nucleases and TALEN nucleases
35 The patent application is set against the backdrop of tools and methodologies used prior to October 2012 for introducing mutations into DNA sequences. The patent application describes how the CRISPR/Cas system is used to recognise and silence exogenous genetic elements in a manner analogous to the process of RNA interference (RNAi) in eukaryotic organisms. RNAi is a biological system in which RNA molecules inhibit gene expression by neutralising targeted mRNA molecules. RNAi was known to those in the field of genetic engineering well before October 2012. The patent application and P1 also refer in particular to the use of other gene-editing tools known as Zinc finger nucleases (ZFNs) and TALEN nucleases (TALENs) that are derived from eukaryotic transcription factors.
36 CRISPR is an acronym for “Clustered Regularly Interspaced Short Palindromic Repeats”. Cas9 is the CRISPR associated protein 9, which is a prokaryotic dual RNA-guided DNA endonuclease (an enzyme that cuts DNA within the internal part of the DNA sequence). These components are associated with the CRISPR/Cas adaptive immune system found in bacteria, an example of which is the Type II CRISPR/Cas system which is characterised by (inter alia) its use of Cas9 protein (or polypeptide). In essence, these systems are a defence mechanism which protects the bacteria from invading viruses by cleaving the DNA of the virus and thereby disabling it.
37 Type II CRISPR/Cas systems were known to exist in certain prokaryotic cells and function in the genomes of bacteria as part of their acquired bacterial immune system. Specifically, Type II CRISPR/Cas systems were understood to confer bacterial resistance to exogenous (external) genetic elements such as plasmids (small circular DNA found in bacteria that replicate in bacterial cells) and phages (viruses that infect and replicate in cells).
38 This bacterial resistance is achieved by way of short segments of plasmid/phage DNA, called spacers, which are incorporated into the bacterial genome between (and separate) CRISPR repeats (short palindromic sequences). Together, these spacers and repeats make up what is known as the CRISPR array. The CRISPR spacers serve as a memory of past exposure to plasmids and phages and are used to recognise and silence foreign DNA from invading bacteria and phages.
39 Figure 2(A) in Horvath (2010) (reproduced below) shows the process of spacer incorporation which is taking DNA from an invading virus or plasmid and incorporating this into the CRISPR array as spacer units. Figure 2(B) illustrates the function of CRISPR-Cas in targeting and cleaving invading DNA that is cognate to a spacer. This is done by transcribing the CRISPR repeat-spacer array into “pre-crRNA” which is then processed through RNA cleavage into individual “crRNA” (CRISPR RNA) units incorporating the spacer and sequence derived from the repeat. The spacer sequence in the crRNA unit guides the crRNA-Cas complex to the invading nucleic acid. The crRNA unit complexed with Cas protein(s) is the active form of the CRISPR defence system which performs the cleavage of the invading DNA.
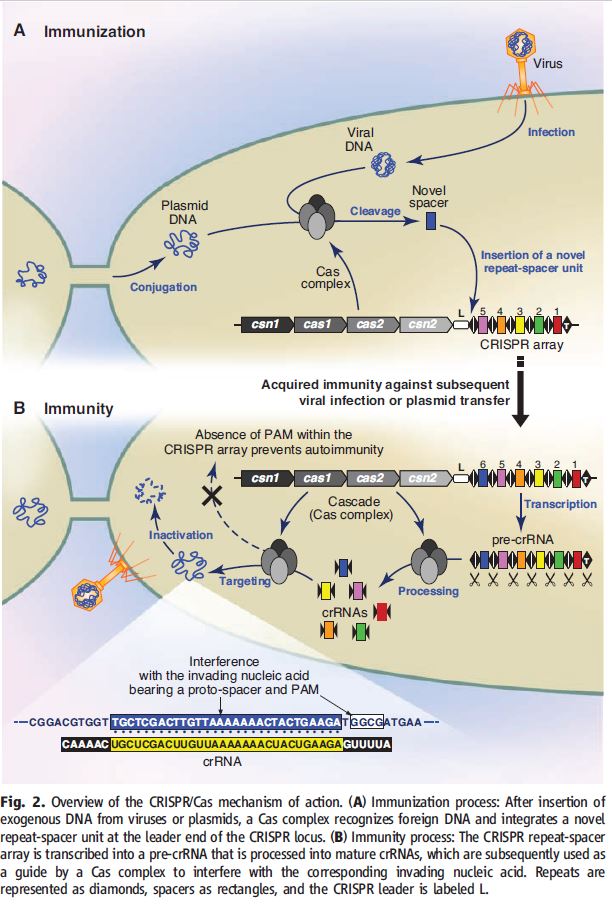
40 The patent application describes how the Cas9 component of the Type II CRISPR/Cas system forms an active endonuclease when complexed with two RNA molecules designated CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA) that are fused together to form a chimeric (single) guide RNA that guide the CRISPR/Cas 9 complex to its target DNA sequence. The RNA guided endonuclease is then able to break foreign genetic elements in invading phages or plasmids and protect the host cell (bacteria) from infection.
41 The composition and role of the crRNA and tracrRNA that make up the single guide RNA (“sgRNA”) are of some importance to understanding the background to the CRISPR/Cas9 system. The crRNA is transcribed from the spacer sequence of the CRISPR array into pre-crRNA which is further processed into mature crRNA that is complementary to the target DNA sequence. This complementarity is used to guide the Cas9 to the target DNA sequence of interest where it will cleave. The tracrRNA is transcribed from a gene outside of the CRISPR array but it is complementary to the repeat sequences of the CRISPR array. This complementarity is used to process pre-crRNA into mature crRNA so that it can perform its guiding function.
42 The invention described in the specification involves the use of individual components of the CRISPR/Cas9 system as a programmable system for introducing a site-specific double-stranded break in a target nucleic acid of a eukaryotic cell (i.e. outside of a prokaryotic system). The system can be used in vitro (outside the cell) or in vivo (in the cell) to introduce mutations into DNA sequences including in so-called “gene-editing” experiments or research.
43 The reference to a site-specific double-stranded break is a reference to the ability of the system to target and cleave a precise site which is within a DNA target sequence that is complementary to the variable part of the guide RNA, and which contains a PAM recognised by Cas9. PAM refers to “protospacer adjacent motif” which is a nucleotide sequence adjacent to the target DNA sequence (known as the protospacer) to be cleaved by Cas9. The PAM sequence is the means by which a Cas9 recognises where to cleave DNA. Each Cas9 derived from a bacterial species recognises a specific PAM sequence. For example, the patent application discloses that Cas9 derived from Streptococcus pyogenes recognises a “NGG” or “NAG” PAM sequence where “N” stands for any nucleotide and “GG” stands for two guanine nucleotides. S. pyogenes Cas9 will therefore cleave DNA adjacent to the nucleotide sequence “NGG” where the Cas9 is complexed with a guide RNA that has a variable region of the crRNA that is complementary to the target DNA sequence.
44 The sgRNA depicted in Figure 1a of the patent application and Figure 1A of P1 are identical and reproduced below. The target DNA sequence is shown in green. The PAM sequence “CGG” recognised by Cas9 is shown in orange and the triangles indicate cleavage sites. Cas9 is shown in yellow and the sequences of the guide RNA derived from crRNA and tracrRNA are shown in red and blue, respectively. Vertical bars between the target DNA and crRNA sequence and between the crRNA and tracrRNA denote complementarity. The coloured boxes do not appear in Figure 1a of the patent application or Figure 1A of P1 and have been added to assist explanation of the figure.
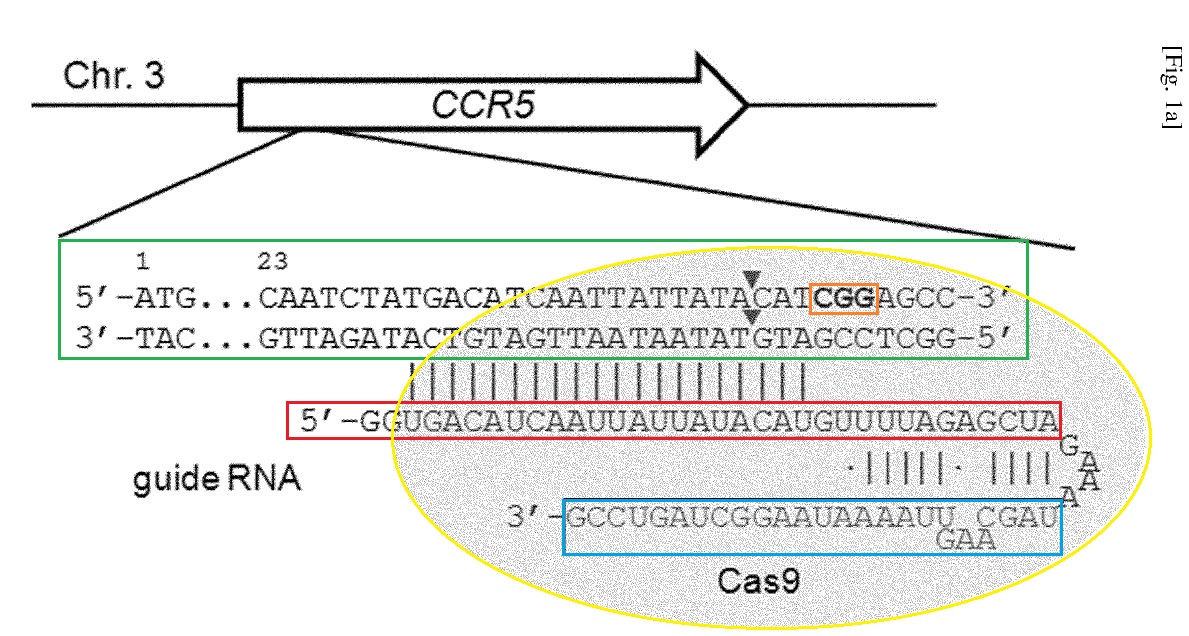
Figure 1A/ Figure 1a
45 In summary, the particular CRISPR/Cas9 system the subject of the patent application (and P1) is said to be comprised of a Cas9 polypeptide, that when complexed with a chimeric (single) guide RNA, has endonuclease activity in eukaryotic cells.
46 There were four principal witnesses each of whom provided written and oral evidence.
Associate Professor Ron Firestein
47 Associate Professor Firestein made two affidavits dated 13 September 2019 (“Firestein 1”) and 25 March 2020 (“Firestein 2”). He also annexed to Firestein 1 his Declaration in the Patent Office opposition proceedings. He is the Head of the Centre for Cancer Research at the Hudson Institute of Medical Research, and a consulting pathologist in molecular genetic pathology at Monash Health. By his expertise and training he can be described as a molecular biologist. At October 2012, Associate Professor Firestein was generally aware of the existence of bacterial innate immunity but did not have knowledge of the mechanistic details of the system, nor was this work of interest to him because the potential impact of the CRISPR/Cas9 system for molecular biology and gene editing had not been identified. He has used the CRISPR/Cas9 system since mid-2013.
48 Professor Giffard made two affidavits dated 13 September 2019 (“Giffard 1”) and 16 March 2020 (“Giffard 2”). He is the Head of Laboratory Science at the Menzies School of Health Research and Professor and Associate Dean for Research and Innovation in the College of Health and Human Sciences at Charles Darwin University. He has specialised knowledge in the field of bacterial genetics and physiology, molecular bacteriology, bioinformatics and molecular microbiology. By his expertise and training he can be described as a microbiologist. Before October 2012, he co-authored two papers reporting research into the CRISPR loci of the bacterial species C. jejuni and the Staphylococcus bacterial strain MSHR1132.
49 Professor Thomas made two affidavits dated 8 April 2019 (“Thomas 1”) and 20 August 2020 (“Thomas 2”). He is a Professor of Biochemistry at the University of Adelaide and is the Head of the Genome Editing Laboratory at the South Australian Health and Medical Research Institute. Since 1995, he has engaged in genetic research in eukaryotes, including gene targeting to inactivate genes of interest using homologous recombination, the development of mouse models with genetic changes, and using molecular biology technology to screen for genetic mutations. By his expertise and training he can be described as a molecular biologist. At October 2012, Professor Thomas was generally aware of the CRISPR/Cas9 system in bacteria, but not to a high level of detail. He did not actively follow literature developments relating to bacterial systems. He became interested in using CRISPR/Cas9 systems in mid-2013 after reading Mali (2013) and Wang (2013). Since then, he has produced more than 60 novel mouse models using CRISPR/Cas9 systems, as well as cell lines with modified genomes.
Associate Professor Marco Herold
50 Associate Professor Herold made two affidavits dated 4 April 2019 (“Herold 1”) and 20 December 2019 (“Herold 2”). He is a molecular biologist and the Laboratory Head at the Walter and Eliza Hall Institute of Medical Research. From 2001, he focused on the molecular regulation of cell death, including by introducing foreign genomic material into the genome of host cells using retroviruses. From 2005, he worked on genetic manipulation using RNA interference (RNAi) technology to silence or “knock down” particular genes in eukaryotes, using both mouse models and in vitro systems. At October 2012, Associate Professor Herold was generally aware of CRISPR/Cas9 systems in bacteria after reading Jinek, but was not aware of the specific details of the system. He started working with CRISPR/Cas9 systems in eukaryotic cells and organisms in May 2013, after reading Cong (2013), Mali (2013) and Wang (2013). Since then, he has made around 220 mouse models using CRISPR/Cas9 systems.
51 There were two expert conclaves held prior to the hearing and two concurrent sessions of expert evidence at the hearing. The first expert conclave included the molecular biologists Associate Professor Firestein, Associate Professor Herold and Professor Thomas who prepared a Joint Expert Report dated 1 September 2020 (“JER 1”). These experts also gave evidence in a concurrent session. The second expert conclave included the microbiologist Professor Giffard, Professor Thomas and Associate Professor Herold. They prepared a Joint Expert Report dated 2 September 2020 (“JER 2”) and also gave evidence in another concurrent session.
THE EARLIEST PRIORITY DOCUMENT (P1)
52 It is common ground that P1 was filed on 23 October 2012. P1 is a relatively short document which resembles an unpublished journal article to which has been added an additional paragraph headed “Summary of the Invention”. Nothing turns on the purpose for which P1 was prepared. P1 states at pages 1-6:
[Page 1]
Abstract:
We present a novel genome editing technology based on RNA-guided Cas9 endonucleases (RGENs). Cas9 is a sequence-specific endonuclease in type II CRISPR/Cas systems, which confer prokaryotes with adaptive immunity against invading phages and plasmids. Cas9 recognizes and cleaves target DNA sequences complementary to small synthetic guide RNAs embedded in this protein, generating site-specific DNA double-strand breaks in vitro and in human cells, whose spontaneous repair induces targeted genome modifications at high frequencies. Unlike ZFNs and TALENs, which are used widely in research and biotechnology, RGENs are customized without any cloning step, making them a broadly useful, scalable and expeditious platform for genome engineering in cells and organisms.
Summary of the Invention
In some embodiments, the present invention provides compositions and methods for research, clinical and screening applications for genome editing. In some embodiments, the present invention provides nucleic acids encoding RNA-guided Cas9 endonucleases, vectors comprising Cas-9 endonucleases, Cas-9 polypeptides, and uses of such compositions.
Additional embodiments are described herein.
[Page 2]
Main Text:
We exploited the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein (Cas) system (1), an adaptive immune response in bacteria and archaea, to develop a novel genome editing technology based on RNA-guided endonucleases (RGENs). Cas9, an essential protein component in the Type II CRISPR/Cas system, forms an active endonuclease when complexed with two RNAs termed CRISPR RNA (crRNA) and transactivating crRNA (tracrRNA), thereby slicing foreign genetic elements in invading phages or plasmids to protect the host cells. crRNA is transcribed from the CRISPR element in the host genome, which was previously captured from such foreign invaders. Recently, Jinek et al. (2) elegantly demonstrated that a single-chain chimeric RNA produced by fusing an essential portion of crRNA and tracrRNA could replace the two RNAs in the Cas9/RNA complex to form a functional endonuclease, raising the possibility of using this system for genome editing in cells and organisms. Here, we present the first evidence that RGENs can indeed induce site-specific genome modifications in mammalian cells at high frequencies.
We first tested the DNA cleavage activity of Cas9 derived from Streptococcus pyogenes in the presence or absence of a chimeric guide RNA in vitro. To this end, we used recombinant Cas9 protein that was expressed in and purified from E. coli to cleave a predigested or circular plasmid DNA that contained the 23-base pair (bp) human CCR5 target sequence. A Cas9 target sequence consists of a 20-bp DNA sequence complementary to crRNA or a chimeric guide RNA and the trinucleotide (5'-NGG-3') protospacer adjacent motif (PAM) recognized by Cas9 itself (Fig. 1A); Cas9 cleaved the plasmid DNA efficiently at the expected position only in the presence of the synthetic RNA and did not cleave a control plasmid that lacked the target sequence (Fig. 1B).
Next, we used a RFP-GFP reporter to investigate whether the Cas9/guide RNA complex can cleave the target sequence incorporated between the RFP and GFP sequences in mammalian [Page 3] cells. In this reporter, the GFP sequence is fused to the RFP sequence out-of-frame (3). The active GFP is expressed only when the target sequence is cleaved by site-specific nucleases, which causes frameshifting small insertions or deletions (indels) around the target sequence via error-prone non-homologous end-joining (NHEJ) repair of the double-strand break (DSB). We co-transfected the Cas9-encoding plasmid, the guide RNA, and the RFP-GFP reporter plasmid into human embryonic kidney (HEK) 293T cells, and found that GFP-expressing cells were obtained only when the cells were co-transfected with the Cas9 plasmid and the guide RNA (Fig. 2), demonstrating that RGENs could recognize and cleave the target DNA sequence in cultured human cells.
To test whether RGENs could be used for targeted disruption of endogenous genes in mammalian cells, we analyzed genomic DNA isolated from transfected cells using T7 endonuclease I (T7E1), a mismatch-sensitive endonuclease that specifically recognizes and cleaves heteroduplexes formed by the hybridization of wild-type and mutant DNA sequences (4). We found that mutations were induced only when the cells were co-transfected with both Cas9 and guide RNA (Fig. 3). Mutation frequencies (Indels (%) in Fig. 3A) estimated from the relative DNA band intensities were RNA-dosage dependent, ranging from 1.3% to 5.1%. DNA sequencing analysis of the PCR amplicons corroborated the induction of RGEN-mediated mutations at the endogenous sites. Indels and microhomologies, characteristic of error-prone NHEJ, were observed at the target site. The mutation frequency measured by direct sequencing was 7.3% (= 7 mutant clones/96 clones), on par with those obtained with zinc finger nucleases (ZFNs) or transcription-activator-like effector nucleases (TALENs).
Both ZFNs and TALENs have been successfully developed to disrupt the human CCR5 gene (4-7), which encodes a G-protein-coupled chemokine receptor, an essential co-receptor of HIV infection. A CCR5-specific ZFN is now under clinical investigation in the US for the treatment of AIDS (8). These ZFNs and TALENs, however, have off-target effects, inducing both local [Page 4] mutations at sites whose sequences are homologous to the on-target sequence (7, 9-11) and genome rearrangements that arise from the repair of two concurrent DSBs induced at on-target and off-target sites (12-13). The most striking off-target sites associated with these CCR5-specific engineered nucleases reside in the CCR2 locus, a close homolog of CCR5, located 15-kbp upstream of CCR5. To avoid off-target mutations in the CCR2 gene and unwanted deletions, inversions, and duplications of the 15-kbp chromosomal segment between the CCR5 on-target and CCR2 off-target sites, we intentionally chose the target site of our CCR5-specific RGEN to recognize a region within the CCR5 sequence that has no apparent homology with the CCR2 sequence.
We investigated whether the CCR5-specific RGEN had off-target effects. To this end, we searched for potential off-target sites in the human genome by identifying sites that are most homologous to the intended 23-bp target sequence. As expected, no such sites were found in the CCR2 gene. Instead, we found four sites, each of which carries 3-base mismatches with the on-target site (Fig. 4A). The T7E1 assays showed that mutations were not detected at these sites (assay sensitivity, ⁓0.5%), demonstrating exquisite specificities of RGENs (Fig. 4B). Furthermore, we used PCR to detect the induction of chromosomal deletions in cells separately transfected with plasmids encoding the ZFN and RGEN specific to CCR5. Whereas the ZFN induced deletions, the RGEN did not (Fig. 4C). Although we did not detect any off-target effects with RGENs in this study, deep sequencing of candidate sites and whole genome or exome sequencing may reveal off-target mutations induced by RGENs.
Next, we reprogrammed RGENs by replacing the CCR5-specific guide RNA with a newlysynthesized RNA designed to target the human C4BPB gene, which encodes the beta chain of C4b-binding protein, a transcription factor. This RGEN induced mutations at the chromosomal target site in K562 cells at high frequencies (Fig. 38): Mutation frequencies measured by the T7E1 assay and by direct sequencing were 14% and 8.3% (= 4 mutant clones/48 clones), [Page 5] respectively. Out of four mutant sequences, two clones contained a single-base or two-base insertion precisely at the cleavage site, a pattern that was also observed at the CCR5 target site. These results indicate that RGENs cleave chromosomal target DNA at expected positions in cells.
ZFNs and TALENs enable targeted mutagenesis in mammalian cells (14-16), model organisms (17-20), plants (21-23), and livestock (24-25), but the mutation frequencies obtained with individual nucleases are widely different from each other. Furthermore, some ZFNs and TALENs fail to show any genome editing activities (26-29). DNA methylation may limit the binding of these engineered nucleases to target sites (30). In addition, it is technically challenging and time-consuming to make custom nucleases. In this regard, RGENs based on Cas9 could provide useful options for genome editing. Compared to ZFNs and TALENs, RGENs can be more readily customized because only the synthetic RNA component is replaced to make a new genome-editing nuclease: No sub-cloning steps are involved to make customized RGENs. Furthermore, the relatively small size of the Cas9 gene (4.2 kbp) as compared to a pair of TALEN genes (⁓6 kbp) provides an advantage for this system in some applications such as virus-mediated gene delivery. These features will make RGENs scalable, versatile, and convenient tools for genome engineering in cells and organisms.
The specificity of DNA recognition by RGENs is somewhat limited by the requirement for a 5'-GG-3' dinucleotide in the PAM sequence. This motif is recognized by the Cas9 protein but not by the guide RNA. Thus, RGENs can be designed to cleave DNA once per 8 bp (= 4x4/2) on average. This limitation might be relieved by engineering Cas9 or employing Cas9 derived from other species.
Unlike Fokl-based ZFNs and TALENs, which produce 4- to 6-base 5' overhangs at cleavage sites, RGENs yield blunt ends rather than cohesive ends (2). Our results show that DSBs with blunt ends can also be readily repaired in mammalian cells. It would be interesting to investigate [Page 6] how and whether blunt DSB ends would be differentially repaired by endogenous end-joining processes.
Taken together, these findings indicate that RGENs are a new member in the family of genome editing tools that have revolutionized basic and biomedical research but with their own unique features that make them an ideal platform in many applications. We propose that RGENs should find broad utility in research, biotechnology, and medicine in the post-genomic era.
53 These three sections of P1 are followed by 30 references to various journal articles, the second of which is the article first published in Science online on 28 June 2012 and published in print on 17 August 2012 by Jinek et al that is of some importance to the issue of enablement. The article is by Martin Jinek, Krzysztof Chylinski, Ines Fonfara, Michael Hauer, Jennifer A. Doudna, and Emmanuelle Charpentier, and is entitled “A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity” Science 337, 816 (2012) (“Jinek”). The question whether the invention involves an inventive step in light of Jinek does not arise in this proceeding.
54 The 30 references are then followed by a description of various figures reproduced in P1 including Figure 1A which includes a schematic representation of a guide RNA and a Cas9 protein used to cleave plasma DNA in vitro. The PAM sequence recognised by the Cas9 protein is identified in Figure 1A as CGG which is a PAM sequence recognised by Cas9 derived from S. pyogenes (ie. a 5’-NGG-3’ PAM where N stands for any nucleotide A, T, C or G). Triangles in Figure 1A depict the site of the intended break which is in a complimentary position on each DNA strand. Figure 3 provides mutation frequencies for RGEN-driven mutations at sites on the CCR5 gene and the C4BPB gene. The authors state that they reprogrammed the guide RNA used with the Cas9 protein to target the CCR5 gene with a different guide RNA to target the C4BPB gene. Accordingly, there is a specific disclosure of the use of two different guide RNAs targeting these two different genes in mammalian cells.
55 The description of the figures is followed by a section of P1 headed “Materials and Methods” which describes (inter alia) the construction of Cas9 encoding plasmids derived from S. pyogenes strain M1 GAS and the preparation of RNA in vitro using a MEGAshortscript T7 kit (Ambion). This is followed by a description of the genome-editing assay whereby mammalian cells (K562 cells) were transfected with 20µg of Cas9-encoding plasmid followed (after 24 hours) by the introduction of 10-40µg of in vitro transcribed chimeric RNA.
56 The complete specification (“the Specification”) is entitled “Composition for cleaving a target DNA comprising a guide RNA specific for the target DNA and Cas protein-encoding nucleic acid or Cas protein, and use thereof”. The Specification is divided into a number of different sections. The first section contains a brief description of the “Technical Field” of the invention. The Specification states at [1]:
The present invention relates to targeted genome editing in eukaryotic cells or organisms. More particularly, the present invention relates to a composition for cleaving a target DNA in eukaryotic cells or organisms comprising a guide RNA specific for the target DNA and Cas protein-encoding nucleic acid or Cas protein, and use thereof.
57 The second section is headed “Background Art” and includes a brief discussion at [3]-[9] of the relevant technology including some prior art. The prior art referred to includes Jinek.
58 The Specification states at [3]-[4]:
[3] CRISPRs (Clustered Regularly Interspaced Short Palindromic Repeats) are loci containing multiple short direct repeats that are found in the genomes of approximately 40% of sequenced bacteria and 90% of sequenced archaea. CRISPR functions as a prokaryotic immune system, in that it confers resistance to exogenous genetic elements such as plasmids and phages. The CRISPR system provides a form of acquired immunity. Short segments of foreign DNA, called spacers, are incorporated into the genome between CRISPR repeats, and serve as a memory of past exposures. CRISPR spacers are then used to recognize and silence exogenous genetic elements in a manner analogous to RNAi in eukaryotic organisms.
[4] Cas9, an essential protein component in the Type II CRISPR/Cas system, forms an active endonuclease when complexed with two RNAs termed CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA), thereby slicing foreign genetic elements in invading phages or plasmids to protect the host cells. crRNA is transcribed from the CRISPR element in the host genome, which was previously captured from such foreign invaders. Recently, Jinek et al. (1) demonstrated that a single-chain chimeric RNA produced by fusing an essential portion of crRNA and tracrRNA could replace the two RNAs in the Cas9/RNA complex to form a functional endonuclease.
59 The Cas9 protein is an essential part of the genome editing technology described. It is an enzyme which, when complexed with a suitable guide RNA, provides a functional endonuclease that cuts each strand of DNA within the internal part of its sequence (rather than at its end) so as to generate a double-stranded break. A nuclease is an enzyme that cleaves DNA. An endonuclease is an enzyme that cuts DNA within the internal part of its sequence (in comparison to an exonuclease which trims the DNA at its ends). Endonucleases can generate two types of “ends”. One type of endonuclease cuts the DNA at the same position on the sense and antisense strands of the DNA to generate what is known as a “blunt” end. Another type of endonuclease cuts DNA at different positions on the sense and antisense strands to generate a “staggered” (or “sticky”) end. This second type of double-stranded break, sometimes referred to as “composite” double-stranded break, will have single stranded overhangs on either the 5’ or 3’ side of the DNA formation.
60 Two well-known gene editing tools discussed in the Specification are Zinc finger nucleases (ZFNs) and TALEN nucleases (TALENs). Zinc finger technology uses artificial restriction enzymes which can be used to cleave (cut) DNA strands generated by fusing a zinc finger DNA-binding domain to a DNA-cleavage domain. Zinc finger nucleases can be engineered to target specific desired DNA sequences and this enables them to target unique sequences within complex genomes. TALEN is an acronym for “Transcription Activator-like Effector Nuclease”, which are DNA-binding proteins that can be engineered to cut specific sequences of DNA. “RFLP” is an acronym for “Restriction Fragment Length Polymorphism”. This refers to the presence of a difference in the nucleotide sequence between two homologous DNA sequences that can be detected by use of a restriction enzyme that will preferentially recognise (and cut) only one variant of those two sequences. RFLP analysis allows polymorphisms to be identified based on the differences in the cutting activity of the restriction enzyme.
61 The Specification states at [5]-[9]:
[5] CRISPR/Cas systems offer an advantage to zinc finger and transcription activator-like effector DNA-binding proteins, as the site specificity in nucleotide binding CRISPR-Cas proteins is governed by a RNA molecule instead of the DNA-binding protein, which can be more challenging to design and synthesize.
[6] However, until now, a genome editing method using the RNA-guided endonuclease (RGEN) based on CRISPR/Cas system has not been developed.
[7] [BLANK]
[8] Meanwhile, Restriction fragment length polymorphism (RFLP) is one of the oldest, most convenient, and least expensive methods of genotyping that is still used widely in molecular biology and genetics but is often limited by the lack of appropriate sites recognized by restriction endonucleases.
[9] Engineered nuclease-induced mutations are detected by various methods, which include mismatch-sensitive T7 endonuclease I (T7El) or Surveyor nuclease assays, RFLP, capillary electrophoresis of fluorescent PCR products, Dideoxy sequencing, and deep sequencing. The T7El and Surveyor assays are widely used but are cumbersome. Furthermore, theses enzymes tend to underestimate mutation frequencies because mutant sequences can form homoduplexes with each other and cannot distinguish homozygous bi-allelic mutant clones from wildtype cells. RFLP is free of these limitations and therefore is a method of choice. Indeed, RFLP was one of the first methods to detect engineered nuclease-mediated mutations in cells and animals. Unfortunately, however, RFLP is limited by the availability of appropriate restriction sites. It is possible that no restriction sites are available at the target site of interest.
62 The “Technical Problem” to which the invention is directed is described as follows at [11]-[13]:
Disclosure of Invention
Technical Problem
[11] Until now, a genome editing and genotyping method using the RNA-guided endonuclease (RGEN) based on CRISPR/Cas system has not been developed.
[12] Under these circumstances, the present inventors have made many efforts to develop a genome editing method based on CRISPR/Cas system and finally established a programmable RNA-guided endonuclease that cleave DNA in a targeted manner in eukaryotic cells and organisms.
[13] In addition, the present inventors have made many efforts to develop a novel method of using RNA-guided endonucleases (RGENs) in RFLP analysis. They have used RGENs to genotype recurrent mutations found in cancer and those induced in cells and organisms by engineered nucleases including RGENs themselves, thereby completing the present invention.
63 The section of the Specification headed “Solution to the Problem” includes at [15] a lengthy statement of what are said to be objects of the invention. At least some of these are in a form that reflects the language of the claims including claims 1 and 10. For convenience, the ten paragraphs that make up [15] have been numbered as [15.1]-[15.10]. All of these paragraphs but for the first appear to have been introduced by amendments made in 2016. The first ten objects are said to be:
[15.1] It is an object of the present invention to provide a composition for cleaving target DNA in eukaryotic cells or organisms comprising a guide RNA specific for target DNA or DNA that encodes the guide RNA, and Cas protein-encoding nucleic acid or Cas protein.
[15.2] It is another object of the present invention to provide a composition for inducing targeted mutagenesis in eukaryotic cells or organisms, comprising a guide RNA specific for target DNA or DNA that encodes the guide RNA, and Cas protein-encoding nucleic acid or Cas protein.
[15.3] It [sic] an object of the present invention to provide a composition comprising a Type II Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas system for use in introducing a site-specific, double stranded break at a target nucleic acid sequence in a eukaryotic cell, said CRISPR/Cas system comprising (i) a nucleic acid encoding a Cas9 polypeptide comprising a nuclear localization sequence, and (ii) a nucleic acid encoding a guide RNA that hybridizes to a target nucleic acid, wherein the guide RNA is a chimeric guide RNA comprising a CRISPR RNA (crRNA) portion fused to a trans activating crRNA (tracrRNA) portion.
[15.4] It is still another object of the present invention to provide a kit for cleaving a target DNA in eukaryotic cells or organisms comprising a guide RNA specific for target DNA or DNA that encodes the guide RNA, and Cas protein-encoding nucleic acid or Cas protein.
[15.5] It is still another object of the present invention to provide a kit for inducing targeted mutagenesis in eukaryotic cells or organisms comprising a guide RNA specific for target DNA or DNA that encodes the guide RNA, and Cas protein-encoding nucleic acid or Cas protein.
[15.6] It is still another object of the present invention to provide a method for preparing a eukaryotic cell or organism comprising Cas protein and a guide RNA comprising a step of co-transfecting or serial-transfecting the eukaryotic cell or organism with a Cas protein-encoding nucleic acid or Cas protein, and a guide RNA or DNA that encodes the guide RNA.
[15.7] It is an objection [sic] of the present invention to provide a method of introducing a site-specific, double-stranded break at a target nucleic acid sequence in a eukaryotic cell, the method comprising introducing into the eukaryotic cell a Type II Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas system, wherein the CRISPR/Cas system comprises:
a) a nucleic acid encoding a Cas9 polypeptide comprising a nuclear localization signal, wherein the nucleic acid is codon-optimized for expression in eukaryotic cells, and
b) a nucleic acid encoding a guide RNA that hybridizes to the target nucleic acid, wherein the guide RNA is a chimeric guide RNA comprising a CRISPR RNA (crRNA) portion fused to a trans activating crRNA (tracrRNA) portion, wherein the target nucleic acid sequence comprises a first strand that binds to the crRNA portion and a second strand having a trinucleotide protospacer adjacent motif (PAM),
and wherein the Cas9 polypeptide and the guide RNA form a Cas9/RNA complex in the eukaryotic cell, whereby a site-specific, double stranded break at the target nucleic acid sequence is introduced.
[15.8] It is still another object of the present invention to provide a eukaryotic cell or organism comprising a guide RNA specific for target DNA or DNA that encodes the guide RNA, and Cas protein-encoding nucleic acid or Cas protein.
[15.9] It is still another object of the present invention to provide a method for cleaving a target DNA in eukaryotic cells or organisms comprising a step of transfecting the eukaryotic cells or organisms comprising a target DNA with a composition comprising a guide RNA specific for target DNA or DNA that encodes the guide RNA, and Cas protein-encoding nucleic acid or Cas protein.
[15.10] It is still another object of the present invention to provide a method for inducing targeted mutagenesis in a eukaryotic cell or organism comprising a step of treating a eukaryotic cell or organism with a composition comprising a guide RNA specific for target DNA or DNA that encodes the guide RNA, and Cas protein-encoding nucleic acid or Cas protein.
(sub-paragraph numbering added)
Mutagenesis refers to changes in DNA (either naturally occurring or artificially engineered) that result in gene mutation.
64 It is apparent that sub-paragraph [15.1] distinguishes between a composition comprising (inter alia) “a guide RNA” that targets a particular DNA sequence and “DNA that encodes the guide RNA”. Sub-paragraph [15.3], which mirrors the language of claim 1, also refers to nucleic acid “encoding” a Cas9 polypeptide and nucleic acid “encoding” a guide RNA. The Specification makes similar use of the word “encoding” at [156] where there is reference to “a component in the form of a protein or in the form of a nucleic acid encoding Cas protein.”
65 Further objects are set out at [31], [33], [35], [37]-[38], [40], [42], [44], [46], [48], [50], [52], [54], [56], [58], [60], [62], [64], [66] and [68]. The advantageous effects of the invention are described as follows at [69]:
The present composition for cleaving a target DNA or inducing a targeted mutagenesis in eukaryotic cells or organisms, comprising a guide RNA specific for the target DNA and Cas protein-encoding nucleic acid or Cas protein, the kit comprising the composition, and the method for inducing targeted mutagenesis provide a new convenient genome editing tools. In addition, because custom RGENs can be designed to target any DNA sequence, almost any single nucleotide polymorphism or small insertion/deletion (indel) can be analyzed via RGEN-mediated RFLP, therefore, the compostion [sic] and method of the present invention may be used in detection and cleaving naturally-occurring variations and mutations.
66 In the section entitled “Best Mode for Carrying out the Invention” the Specification states at [137] – [145]:
[137] In accordance with one aspect of the invention, the present invention provides a composition for cleaving target DNA in eukaryotic cells or organisms comprising a guide RNA specific for target DNA or DNA that encodes the guide RNA, and Cas protein-encoding nucleic acid or Cas protein. In addition, the present invention provides a use of the composition for cleaving target DNA in eukaryotic cells or organisms comprising a guide RNA specific for target DNA or DNA that encodes the guide RNA, and Cas protein-encoding nucleic acid or Cas protein.
[138] [BLANK]
[139] In the present invention, the composition is also referred to as a RNA-guided endonuclease (RGEN) composition.
[140] [BLANK]
[141] ZFNs and TALENs enable targeted mutagenesis in mammalian cells, model organisms, plants, and livestock, but the mutation frequencies obtained with individual nucleases are widely different from each other. Furthermore, some ZFNs and TALENs fail to show any genome editing activities. DNA methylation may limit the binding of these engineered nucleases to target sites. In addition, it is technically challenging and time-consuming to make customized nucleases.
[142] [BLANK]
[143] The present inventors have developed a new RNA-guided endonuclease composition based on Cas protein to overcome the disadvantages of ZFN s and TALENs.
[144] [BLANK]
[145] Prior to the present invention, an endonuclease activity of Cas proteins has been known. However, it has not been known whether the endonuclease activity of Cas protein would function in an eukaryotic cell because of the complexity of the eukaryotic genome. Further, until now, a composition comprising Cas protein or Cas protein-encoding nucleic acid and a guide RNA specific for the target DNA to cleave a target DNA in eukaryotic cells or organisms has not been developed.
67 The Specification refers at [153] to the three types of CRISPR-Cas system found in bacteria including the Type II system involving the Cas9 protein. However, as will been seen, the claims all involve compositions or methods that make use of the Type II system and the Cas9 protein which is integral to that system.
68 According to the Specification at [158]-[159], in the present invention, the Cas protein may be any Cas protein provided that it has an endonuclease or nickase activity when complexed with a guide RNA, but preferably, it is Cas9 protein or variants thereof. The Specification states at [161]-[162]:
[161] Further, Cas protein may be the one isolated from an organism such as Streptococcus sp., preferably Streptococcus pyogens or a recombinant protein, but it is not limited thereto.
[162] The Cas protein derived from Streptococcus pyogens may recognizes NGG trinucleotide. The Cas protein may comprise an amino acid sequence of SEQ ID NO: 109, but it is not limited thereto.
(Errors in original).
69 Streptococcus pyogenes (“S. pyogenes”) is a particular species of bacteria which recognises the PAM sequence “NGG” (5’-NGG-3’ PAM) where N designates any nucleotide and “GG” represents two guanine nucleotides running in the 5’ to 3’ direction.
70 The Specification also states at [176]-[179]:
[176] The guide RNA may be transferred into a cell or an organism in the form of RNA or DNA that encodes the guide RNA. The guide RNA may be in the form of an isolated RNA, RNA incorporated into a viral vector, or is encoded in a vector. Preferably, the vector may be a viral vector, plasmid vector, or agrobacterium vector, but it is not limited thereto.
[177] A DNA that encodes the guide RNA may be a vector comprising a sequence coding for the guide RNA. For example, the guide RNA may be transferred into a cell or organism by transfecting the cell or organism with the isolated guide RNA or plasmid DNA comprising a sequence coding for the guide RNA and a promoter.
[178] Alternatively, the guide RNA may be transferred into a cell or organism using virus-mediated gene delivery.
[179] When the guide RNA is transfected in the form of an isolated RNA into a cell or organism, the guide RNA may be prepared by in vitro transcription using any in vitro transcription system known in the art. The guide RNA is preferably transferred to a cell in the form of isolated RNA rather than in the form of plasmid comprising encoding sequence for a guide RNA. As used herein, the term “isolated RNA” may be interchangeable to “naked RNA”. This is cost- and time-saving because it does not require a step of cloning. However, the use of plasmid DNA or virus-mediated gene delivery for transfection of the guide RNA is not excluded.
71 Transfection refers to the introduction of foreign DNA into a cell. The first sentence of [176] indicates that the guide RNA may take the form of isolated (or naked) RNA introduced into the cell or, alternatively, guide RNA encoded by DNA in the cell. The DNA may be transfected (introduced) into the cell using a vector (i.e. a plasmid). Once inside the cell, the transcription process will be initiated by a promoter included in the plasmid and the DNA will then transcribe the guide RNA. Thus, as [179] indicates, the guide RNA can be prepared in vitro before it is introduced into the cell in the form of “isolated” or “naked” RNA, or the guide RNA can be prepared in vivo after a plasmid containing the RNA-encoding DNA is transfected into the cell. The use of isolated RNA prepared in vitro is said to be preferable to the use of RNA-encoding plasmid DNA prepared in vivo because the former is the cheaper and less time consuming alternative and it does not involve a cloning step of inserting a target DNA fragment into a plasmid.
72 A nickase is an enzyme that cuts only one of the two strands of DNA to create a single strand break in the DNA. A Cas9 nickase is a mutant version of the wild-type Cas9 protein. A “paired Cas nickase” as defined in the Specification “may refer to the guide RNA and the Cas protein functioning as a pair” which may be used to make two breaks at the same or different locations on each of the complementary DNA strands. The Specification suggests that there may be advantages in using paired Cas9 nickases. The discussion in the Specification concerning Example 7 suggests that paired Cas9 nickases may produce composite double-stranded breaks which trigger DNA repair leading to efficient mutagenesis (ie. the generation of mutations) and a doubling in the specificity of Cas9-based genome editing. Various other possible advantages are also discussed.
73 Claims 1 to 21 are as follows:
1 A composition comprising a Type II Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas system for use in introducing a site-specific, double stranded break at a target nucleic acid sequence in a eukaryotic cell, said CRISPR/Cas system comprising (i) a nucleic acid encoding a Cas9 polypeptide comprising a nuclear localization sequence, and (ii) a nucleic acid encoding a guide RNA that hybridizes to a target nucleic acid, wherein the guide RNA is a chimeric guide RNA comprising a CRISPR RNA (crRNA) portion fused to a trans activating crRNA (tracrRNA) portion.
2. The composition of claim 1, wherein said Cas9 polypeptide is a Streptococcus Cas9 polypeptide.
3. The composition of claim 2, wherein said Cas9 polypeptide is a Streptococcus pyogenes Cas9 polypeptide.
4. The composition of any one of claims 1-3, wherein said nucleic acid encoding a Cas9 polypeptide is codon-optimized for expression in eukaryotic cells.
5. The composition of claim 4, wherein said nucleic acid encoding a Cas9 polypeptide is codon-optimized for expression in mammalian cells.
6. The composition of any one of claims 1-5, wherein said nuclear localization sequence is located at the C terminus of the Cas9 polypeptide.
7. The composition of any one of claims 1-5, wherein the target nucleic acid is an endogenous target nucleic acid.
8. The composition of any one of claims 1-5, wherein the guide RNA is in the form of a vector.
9. The composition of any one of claims 1-5, wherein said guide RNA comprises 2 additional guanine nucleotides at the 5' end.
10. A method of introducing a site-specific, double-stranded break at a target nucleic acid sequence in a eukaryotic cell, the method comprising introducing into the eukaryotic cell a Type II Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas system, wherein the CRISPR/Cas system comprises:
(a) a nucleic acid encoding a Cas9 polypeptide comprising a nuclear localization signal, wherein the nucleic acid is codon-optimized for expression in eukaryotic cells, and
(b) a nucleic acid encoding a guide RNA that hybridizes to the target nucleic acid, wherein the guide RNA is a chimeric guide RNA comprising a CRISPR RNA (crRNA) portion fused to a trans activating crRNA (tracrRNA) portion, wherein the target nucleic acid sequence comprises a first strand that binds to the crRNA portion and a second strand having a trinucleotide protospacer adjacent motif (PAM),
and wherein the Cas9 polypeptide and the guide RNA form a Cas9/RNA complex in the eukaryotic cell, whereby a site-specific, double stranded break at the target nucleic acid sequence is introduced.
11. The method of claim 10, wherein the Cas9 polypeptide is a Streptococcus Cas9 polypeptide.
12. The method of claim 11, wherein the Cas9 polypeptide is a Streptococcus pyogenes Cas9 polypeptide.
13. The method of any one of claims 10-12, wherein the nucleic acid encoding the Cas 9 polypeptide is codon-optimized for expression in mammalian cells.
14. The method of any one of claims 10-13, wherein the nuclear localization signal is located at the C terminus of the Cas9 polypeptide.
15. The method of any one of claims 10-14, wherein the eukaryotic cell is a mammalian cell.
16. The method of claim 15, wherein the mammalian cell is a human cell.
17. The method of any one of claims 10-16, wherein the target nucleic acid sequence is a genomic sequence located at its endogenous site in the genome of the eukaryotic cell.
18. The method of any one of claims 10-16, wherein the nucleic acid encoding the guide RNA is a vector.
19. The method of any one of claims 10-16, wherein the nucleic acid encoding the guide RNA is in vitro transcribed RNA.
20. The method of any one of claims 10-16, wherein said guide RNA comprises 2 additional guanine nucleotides at the 5' end.
21. The method of any one of claims 10-16, wherein the nucleic acid encoding the Cas9 polypeptide is introduced into the eukaryotic cell before introducing the nucleic acid encoding the guide RNA into the eukaryotic cell.
74 The claims refer to a “Cas9 polypeptide” rather than a “Cas9 protein” but, as used in both the description of the invention and the claims, these terms have the same meaning and are used interchangeably.
75 The following points should also be noted:
(1) Each of the independent claims 1 and 10 also refer to “nucleic acid encoding” a chimeric guide RNA. There is a question as to whether these words encompass a guide RNA that is prepared in vitro (outside the cell) and introduced into the cell in naked (or isolated) form or whether the claim limits what is described to guide RNA encoded by DNA in vivo (in the cell).
(2) Each of the independent claims 1 and 10 also refer to a composition or method for introducing “a site-specific, double-stranded break”. There is a question as to whether these words are apt to describe not only a blunt end break made by a single active endonuclease, but also a break having staggered ends of the kind made using “paired Cas nickase” in which there will be two Cas9 polypeptides each with its own guide RNA and each producing its own single strand break.
(3) Neither of the independent claims is limited to a Cas9 polypeptide derived from S. pyogenes (although some dependent claims are) that recognises the 5’-NGG-3’ PAM.
(4) Claim 8 refers to the relevant composition wherein the guide RNA “is in the form of a vector”. It is common ground, and I accept, that this should be understood as guide RNA encoded by DNA in a vector.
THE NOTIONAL SKILLED ADDRESSEE
76 The question of who is the notional skilled addressee (or person skilled in the art) arises both in relation to P1 and the patent application.
77 There was a dispute between the parties as to the identity of the notional skilled addressee both in relation to the patent application and P1. ToolGen contends that the notional skilled addressee comprises a team that includes a molecular biologist such as Associate Professors Firestein and Herold and Professor Thomas and a microbiologist such as Professor Giffard. The respondents says that the skilled team does not include a microbiologist. On that basis they contend that Professor Giffard’s evidence is not relevant.
78 The notional skilled addressee is a legal construct and a tool of analysis framed by reference to the available evidence. This will include the patent specification and, typically, evidence of persons with knowledge and experience in the field of the invention.
79 The notional skilled addressee is a person who is likely to have a practical interest in the subject matter of the invention: Catnic Components Ltd v Hill & Smith Ltd [1982] RPC 183 at 242 per Lord Diplock. A person may have a practical interest in an invention at a number of levels. He or she may have an interest in using the products or methods of the invention, making the products of the invention, or making products used to carry out the methods of the invention either alone or in collaboration with others having such an interest: Apotex Pty Ltd v Warner-Lambert Company LLC (No 2) (2016) 122 IPR 17 (“Warner-Lambert”) at [27]. Broadly speaking, the skilled addressee will be a person who also has knowledge and experience in the field of the invention and who will bring to the reading of the relevant document the background knowledge and experience available to those working in that field.
80 In General Tire & Rubber Company v Firestone Tyre & Rubber Company Limited [1972] RPC 457 (“General Tire”) the English Court of Appeal referring to both the construction of the patent in suit and relevant prior art said at 485:
… If the art is one having a highly developed technology, the notional skilled reader to whom the document is addressed may not be a single person but a team, whose combined skills would normally be employed in that art in interpreting and carrying into effect instructions such as those which are contained in the document to be construed. We have already described the composite entity deemed to constitute the notional skilled addressee.
81 In some cases involving complex technology in which the notional skilled addressee is a team, the composition of the team may vary depending on the issue under consideration. As observed by Jacob LJ in Schlumberger Holdings Ltd v Electromagnetic Geoservices AS [2010] RPC 33 at [44] “… the notional team for considering obviousness may have wider skills than the team required for sufficiency” (original emphasis). Referring to Genentech Inc’s Patent [1989] RPC 147, his Lordship observed at [45]:
On the facts the patent was held obvious. The important point to note for present purposes is that the team for obviousness included a protein chemist whereas the team for implementation (sufficiency) did not need him. Different teams for different purposes.
82 The invention described in P1 is said to be a novel genome editing technology. The system described is said to be based on RNA-guided endonucleases (RGENs). The RGENs described in P1 use a Type II CRISPR/Cas system in which the Cas9 protein, when complexed with a guide RNA (crRNA) and trans-activating RNA (tracrRNA), forms an active endonuclease. It is apparent from the opening paragraphs of P1 that it follows on from Jinek which P1 describes as raising the possibility of using the system disclosed in that publication for genome editing in cells and organisms. It is clear from P1 that the focus of the inventors was on the use of their invention in genome editing in eukaryotic cells and in human cells in particular.
83 In support of its submission that the notional skilled addressee would comprise a team including a microbiologist, ToolGen relied on the reference to Jinek in P1 and evidence given by Associate Professor Firestein that the reference to Jinek was directing him to an important paper regarding the development and repurposing of CRISPR/Cas9 technology.
84 I do not consider that P1 directs the notional skilled addressee to Jinek at least not as a source of anything more than general background that gives context to the invention described in P1.
85 It was submitted by ToolGen that the reference to Jinek in P1 must be regarded as part of the disclosure of P1, because the “draftsman had adopted the cross-referencing system solely as a shorthand means of incorporating a writing disclosing the invention”. The authority relied upon by ToolGen in support of that proposition is a passage in the judgment of Lockhart J in Nicaro Holdings Pty Ltd v Martin Engineering Co (1990) 16 IPR 545. In the context of considering whether a prior publication disclosed all the features of the invention of the patent in suit, his Honour said at 549:
The invention must appear in a single disclosure, so it is not permissible to make a pattern or mosaic of or to read together various pieces of prior art in different patents. It is however, permissible, to refer not only to the patent relied on as the source of disclosure but to another patent or other patents incorporated by reference provided that it is plain that the incorporation by reference unequivocally and plainly demonstrates that the draftsman has adopted the cross-referencing system solely as a shorthand means of incorporating a writing disclosing the invention: George C Warner Laboratories Pty Ltd v Chemspray Pty Ltd (1967) 37 AOJP 2513 at 2516; Blanco White, 5th ed, at para 4.107 and Gratwick, “Having Regard to What was Known and Used” (1972) 88 LQR 341 at 343.
86 In my view, the reference to Jinek in P1 falls well short of meeting that test.
87 The evidence of the molecular biologists made clear that the invention disclosed in P1 could be performed without recourse to Jinek. However, ToolGen contended that the notional skilled team (which on its case includes a microbiologist) would refer to Jinek for the purpose of producing a RGEN derived from a bacterial species other than S. pyogenes. The difficulty with this argument is that the reference in P1 to “Cas9 derived from other species” is a mere conjecture that does not form part of the invention described in P1. Importantly, the inventors do not state that other species can be used to perform the invention. The suggestion is much more tentative and does no more than refer to the possibility that other species might prove useful in overcoming the need for a 5’-GG-3’ dinucleotide in the PAM sequence. The fact that a microbiologist with expertise in bacterial CRISPR/Cas systems might be engaged by a molecular biologist who was interested in exploring that possibility is in my opinion not of itself sufficient to justify a finding that such a person would be part of the skilled team. In any event, all of the molecular biologists said that they would not have sought to establish if Cas9 from bacteria other than S. pyogenes recognises a non-NGG PAM on reading P1.
88 In my opinion, the notional skilled addressee to whom P1 is addressed is a molecular biologist with expertise in the field of gene editing in eukaryotic cells. The reference to Jinek in P1 does not justify a finding that a microbiologist would be part of the skilled team.
89 The molecular biologist (working alone or with other molecular biologists and laboratory assistants) will be a highly qualified scientist with a PhD in the field of molecular biology with expertise in gene screening, targeting and manipulation in eukaryotes including in mouse models and in vitro systems. They will be engaged in high level research in a well-resourced laboratory in a medical research institute.
90 If, contrary to my finding, there was a microbiologist on the team, they would have a PhD in the field of microbiology or bacteriology with expertise in the CRISPR/Cas systems found in bacteria and archaea. This member of the team would most likely be engaged in academic research within the biology faculty or department of a University.
The Patent Application
91 In the present case, ToolGen submitted that the patent application is directed to the repurposing of bacterial Type II CRISPR/Cas systems for cleaving target DNA in eukaryotic cells or organisms and that the skilled addressee will comprise a team that includes a person with knowledge of these systems. Hence, ToolGen says that the notional team will have expertise in the field of gene editing in eukaryotic cells, and the expertise of a microbiologist with expertise in bacterial CRISPR/Cas systems. It is on this basis that ToolGen contends that the team will include persons with the knowledge and experience of Associate Professors Herold and Firestein and Professor Thomas (as experts in gene editing in eukaryotic cells) and also a person like Professor Giffard (an expert in bacterial CRISPR/Cas systems). In support of its submissions, ToolGen refers to the description in the patent application of Type II CRISPR/Cas systems involving Cas9 and crRNA and tracrRNA and, in particular, the statements to the effect that the invention is not limited to such systems from S. pyogenes.
92 The respondents submitted that the person who has a practical interest in the subject matter of the invention is a molecular biologist with an interest in genome editing in eukaryotic cells. They submitted that this reflects the “field of the invention” specified in the patent application as well as the express purpose of the invention as described and claimed. On that basis they submitted that a microbiologist such as Professor Giffard, with expertise in relation to bacteria and prokaryotes (not eukaryotic cells) would not form part of the team. They submitted that a microbiologist would have no interest in the subject matter of the invention as described and claimed and that such a person would not be interested in making or using the compositions or methods of the claims for genome editing in eukaryotic cells.
93 The invention disclosed in the patent application is not limited to a system which uses a Cas9 protein derived from S. pyogenes. The Specification makes clear at [161]-[162] that the Cas9 protein which is integral to the Type II CRISPR/Cas system may be derived from S. pyogenes but is not limited to that bacteria and that it may be derived from a different organism. The claims also make clear that claims 1 and 10 are not limited to Cas9 derived from S. pyogenes and that they may be derived from other species. I think it must follow that the invention, as described and claimed, is directed to a notional team including a person with expertise in identifying Cas9 derived from other bacterial species.
94 The respondents submitted that the notional skilled addressee should not be defined in order to fill a gap in the disclosure of the patent application. It submitted that it was not legitimate to attempt to make up for an absence of disclosure by artificially adding to the skilled team a person whose skills might be thought to assist with the development of products or methods that are not disclosed.
95 As I have explained, the patent application makes clear that the invention can be performed using different species of bacteria. Unlike P1, the statements to that effect are clear and unequivocal and indicate that the invention extends to systems that use Cas9 derived from other bacterial species. Molecular biologists reading the patent application would necessarily understand that the invention is not confined to a system that uses Cas9 derived from one species only. If they wished to perform the invention using Cas9 derived from a different bacterial species they would turn to a microbiologist with expertise in CRISPR/Cas systems who could assist in identifying a suitable substitute for S. pyogenes. For that reason I think such a person should be taken to form part of the notional skilled team.
96 In my opinion, the notional skilled addressee to whom the patent application is addressed is a team comprising a molecular biologist with expertise in the field of gene editing in eukaryotic cells and a microbiologist with expertise in CRISPR/Cas systems.
97 The common general knowledge is a general body of knowledge attributed to the hypothetical non-inventive skilled person or team. In Minnesota Mining and Manufacturing Company v Beiersdorf (Australia) Limited (1980) 144 CLR 253, Aickin J at 292 referred to common general knowledge as “… the background knowledge and experience which is available to all in the trade in considering the making of new products, or the making of improvements in old, and it must be treated as being used by an individual as a general body of knowledge”. It is knowledge actually known and used by skilled persons in the relevant field generally or accepted by the bulk of those who are engaged in the particular art: Ranbaxy Laboratories Ltd v AstraZeneca AB (2013) 101 IPR 11, Middleton J at [215] (“Ranbaxy Laboratories”) citing British Acoustic Films Ltd v Nettlefold Productions (1936) 53 RPC 221 at 250 (“British Acoustic Films”). Knowledge is not common general knowledge unless it is sufficiently widely known or used to become generally accepted and assimilated into the minds of people skilled in the relevant art and therefore part of the common general knowledge: see Gilead Sciences Pty Ltd v Idenix Pharmaceuticals LLC (2016) 117 IPR 252 at [210]-[214] (“Gilead”) where Jagot J referred to some of the leading authorities including, Aktiebolaget Hässle v Alphapharm Pty Ltd (2002) 212 CLR 411 at [31]. It will include publications to which the skilled addressee would refer as a matter of course: ICI Chemicals and Polymers Ltd v Lubrizol Corporation Inc (1999) 45 IPR 577 at [112] per Emmett J.
98 However, information does not become common general knowledge merely because it might appear in a journal even if it is one read by persons in the art: Ranbaxy Laboratories at [217] per Middleton J citing (inter alia) Eli Lilly & Co Ltd v Apotex Pty Ltd (2013) 100 IPR 451 at [468]; Wake Forest University Health Sciences v Smith & Nephew Pty Ltd (No 2) (2011) 92 IPR 496 at [96], General Tire at 482-3. In the latter case, the English Court of Appeal referred with approval to the following passage in the judgment of Luxmoore J in British Acoustic Films at 250:
In my judgment it is not sufficient to prove common general knowledge that a particular disclosure is made in an article, or series of articles, in a scientific journal, no matter how wide the circulation of that journal may be, in the absence of any evidence that the disclosure is accepted generally by those who are engaged in the art to which the disclosure relates. A piece of particular knowledge as disclosed in a scientific paper does not become common general knowledge merely because it is widely read, and still less because it is widely circulated. Such a piece of knowledge only becomes general knowledge when it is generally known and accepted without question by the bulk of those who are engaged in the particular art; in other words, when it becomes part of their common stock of knowledge relating to the art.
99 I have applied these principles in this case in determining the common general knowledge at the priority date. In that regard, I accept that certain publications relied on by the appellants (Horvath (2010), Bhaya (2011), Deltcheva (2011) and Makarova (2011)) were common general knowledge to a microbiologist specialising in the CRISPR/Cas systems of prokaryotes at the priority date and therefore common general knowledge of the skilled team if it included a microbiologist working in that field. Further, the matters referred to in [20]-[35] above were common general knowledge of a molecular biologist with expertise in the field of gene editing in eukaryotic cells at the priority date. The matters referred to in [36]-[39] above were common general knowledge of a microbiologist with expertise in CRISPR/Cas systems of prokaryotes at the priority date.
100 Jinek was first published online in Science on 28 June 2012, and in print on 17 August 2012, and is referred to in the patent application and P1. I am not persuaded that Jinek was common general knowledge at the priority date. Associate Professor Herold gave evidence that he had read Jinek before the priority date. Associate Professor Firestein gave evidence that it was very likely he may have read the title and abstract of Jinek but that he had not read the paper in full before the priority date. Professor Thomas gave evidence that he had not read Jinek as at the priority date. There was no evidence led from Professor Giffard, (assuming he is to be treated as a member of the skilled team) that he had read or had even heard of Jinek before the priority date. The evidence does not show that Jinek was widely known and accepted as at the priority date.
101 There was no dispute between the parties as to the principles governing the construction of a patent specification or, in this case, a patent application. The relevant principals were conveniently summarised by the Full Court in Jupiters Ltd v Neurizon Pty Ltd (2005) 222 ALR 155 (Hill, Finn and Gyles JJ) as follows at [67]:
…
(i) the proper construction of a specification is a matter of law: Décor Corporation Pty Ltd v Dart Industries Inc (1988) 13 IPR 385 at 400;
(ii) a patent specification should be given a purposive, not a purely literal, construction: Flexible Steel Lacing Co v Beltreco Ltd (2000) 49 IPR 331; [2000] FCA 890 at [81] (Flexible Steel Lacing); and it is not to be read in the abstract but is to be construed in the light of the common general knowledge and the art before the priority date: Kimberley-Clark Australia Pty Ltd v Arico Trading International Pty Ltd (2001) 207 CLR 1; 177 ALR 460; 50 IPR 513; [2001] HCA 8 at [24];
(iii) the words used in a specification are to be given the meaning which the normal person skilled in the art would attach to them, having regard to his or her own general knowledge and to what is disclosed in the body of the specification: Décor Corporation Pty Ltd at 391;
(iv) while the claims are to be construed in the context of the specification as a whole, it is not legitimate to narrow or expand the boundaries of monopoly as fixed by the words of a claim by adding to those words glosses drawn from other parts of the specification, although terms in the claim which are unclear may be defined by reference to the body of the specification: Kimberley-Clark v Arico at [15]; Welch Perrin & Co Pty Ltd v Worrel (1961) 106 CLR 588 at 610; Interlego AG v Toltoys Pty Ltd (1973) 130 CLR 461 at 478; the body of a specification cannot be used to change a clear claim for one subject matter into a claim for another and different subject matter: Electric & Musical Industries Ltd v Lissen Ltd [1938] 4 All ER 221 at 224–5; (1938) 56 RPC 23 at 39;
(v) experts can give evidence on the meaning which those skilled in the art would give to technical or scientific terms and phrases and on unusual or special meanings to be given by skilled addressees to words which might otherwise bear their ordinary meaning: Sartas No 1 Pty Ltd v Koukourou & Partners Pty Ltd (1994) 30 IPR 479 at 485–6 (Sartas No 1 Pty Ltd); the court is to place itself in the position of some person acquainted with the surrounding circumstances as to the state of the art and manufacture at the time (Kimberley-Clark v Arico at [24]); and
(vi) it is for the court, not for any witness however expert, to construe the specification; Sartas No 1 Pty Ltd at 485–6.
102 The notion of purposive construction, as explained by the Full Court in GlaxoSmithKline Consumer Healthcare Investments (Ireland) (No 2) Limited v Generic Partners Pty Limited (2018) 264 FCR 474 (“GlaxoSmithKline”), requires that the specification be read as a whole and in light of the common general knowledge, and that a practical and common sense approach be adopted to construction. The Full Court said at [106]-[110]:
[106] More recent cases have continued to emphasise the need to read a patent specification as a whole and in light of the common general knowledge. They also confirm that a patent specification should be read in a practical and common sense way and given a “purposive” construction. This approach to construction requires the court to read the specification through the eyes of the skilled addressee with practical knowledge and experience in the field of work in which the invention was intended to be used and a proper understanding of the purpose of the invention.
[107] A well-known case in which these principles were applied was the earlier decision of the House of Lords in Catnic Components Ltd v Hill & Smith Ltd [1982] RPC 183 (HL). The question in that case was whether the defendant had infringed the plaintiffs’ patent for a galvanized steel lintel that the claim required should have a rear support member “extending vertically” from the base plate. The defendant’s lintel was not precisely vertical, but instead extended upwardly at an angle of 84°. The House of Lords held that the defendant’s lintel was nevertheless within the claim. This was because the person skilled in the art would recognise that in order to perform the same function as the rear support member described in the claim, it was not essential that the rear support member in the defendant’s lintel be precisely vertical.
[108] One criticism made of the decision of the House of Lords in Catnic was that it permitted the court to adopt an interpretation of a claim that travelled beyond what was conveyed by the language used so that the forbidden territory extended to devices with rear support members that were not truly vertical. However, as Lord Hoffmann later explained in Kirin-Amgen Inc v Hoechst Marion Roussel Ltd (2004) 64 IPR 444 at [34]:
“Purposive construction” does not mean that one is extending or going beyond the definition of the technical matter for which the patentee seeks protection in the claims. The question is always what the person skilled in the art would have understood the patentee to be using the language of the claim to mean. And for this purpose, the language he has chosen is usually of critical importance. The conventions of word meaning and syntax enable us to express our meanings with great accuracy and subtlety and the skilled man will ordinarily assume that the patentee has chosen his language accordingly. As a number of judges have pointed out, the specification is a unilateral document in words of the patentee’s own choosing. Furthermore, the words will usually have been chosen upon skilled advice. The specification is not a document inter rusticos for which broad allowances must be made. On the other hand, it must be recognised that the patentee is trying to describe something which, at any rate in his opinion, is new; which has not existed before and of which there may be no generally accepted definition. There will be occasions upon which it will be obvious to the skilled man that the patentee must in some respect have departed from conventional use of language or included in his description of the invention some element which he did not mean to be essential. But one would not expect that to happen very often.
[109] It is important to note that Lord Hoffmann was referring here to the meaning conveyed to the skilled addressee by the language used and was not directing himself to a situation in which the skilled addressee deduced that the language of the claim, although conveying to him or her a particular meaning, could never have been intended to mean what it conveyed.
[110] Thus, a skilled addressee may understand a claim that required that something be “vertical” to mean “substantially vertical” or that a claim that includes a requirement that a device “prevents air from coming into contact with the surface of the ink” did not require that it prevent all the air from doing so: Henrikson v Tallon [1965] RPC 434 (HL). These are situations in which the court endeavours to give effect to the skilled addressee’s understanding of the claim language in preference to a purely literal or grammatical construction, not because the skilled addressee understands that the claim contains a mistake that requires correction, but because, when read in the context of the document as a whole and the common general knowledge, the words used would convey that meaning to the skilled addressee.
103 The speech of Lord Hoffman in Kirin-Amgen Inc v Hoechst Marion Roussel Ltd (2004) 64 IPR 444 (“Kirin-Amgen”) has been referred to with approval in many decisions Full Court decisions. In Kimberly-Clark Australia Pty Limited v Multigate Medical Products Pty Limited (2011) 92 IPR 21 (“Multigate”), which was concerned with s 40 of the Act prior to its amendment by the RTB Act, the plurality (Greenwood and Nicholas JJ) said at [44]-[45]:
[44] Since a patent specification must include a detailed description of the invention, and the best method known to the inventor of performing the invention, a patent specification will usually contain a detailed description of at least one embodiment of the invention. But provided a claim is fairly based upon matter disclosed in the specification, a claim may define the invention as having fewer integers than are present in an embodiment so described. Equally, the claim may define an invention as having more features than are present in any such embodiment. It is always open to the patentee to introduce such a limitation if he or she so chooses provided that the claim is fairly based on the matter disclosed in the specification. But if such a limitation has been introduced, it cannot be disregarded simply because the patentee could have framed a valid claim without it.
[45] A patentee may have good reason for introducing a limitation into a claim. As Lord Hoffman explained in Kirin-Amgen Inc v Hoechst Marion Roussel Ltd (2004) 64 IPR 444; [2005] 1 All ER 667; [2004] UKHL 46 at [35] (Kirin-Amgen), a seemingly inexplicable limitation may have been introduced to avoid arguments in relation to prior art. Lord Upjohn made the same point in Rodi & Wienenberger AG v Henry Showell Ltd [1969] RPC 367 at 392 (Rodi): “… some claims may on a superficial reading appear to be unnecessarily circumscribed, but those who have drafted them may have done so in the light of the prior art …”. See also the judgment of Dixon J in Walker v Alemite Corporation (1933) 49 CLR 643 at 656; [1933] ALR 437 (Walker) where his Honour quoted the following well known statement of Lord Parker in Fellows v Thomas William Lench Ltd (1917) 34 RPC 45 at 55 who said “[a] claiming clause operates as a disclaimer of what is not specifically claimed and for such disclaimer there may be reasons known to the inventor but not to the court”.
104 Their Honours also said at [41]:
[41] There are two aspects to the principle which requires that a patent specification be given a purposive rather than a purely literal construction. The first concerns the well recognised need to read words in their proper context. The second is directly related to the nature and function of a patent specification. It is a document that is taken as intended to be read through the eyes of the skilled addressee who is equipped with the common general knowledge in the relevant art. The question is what, in an objective sense, such a person would understand the relevant words of the claim to mean. Ultimately, however, it is the claim that must be construed, and it is not permissible to vary or qualify the plain and unambiguous meaning of the claim by reference to the body of the specification: Interlego AG v Toltoys Pty Ltd (1973) 130 CLR 461 at 478.
105 GlaxoSmithKline was a case in which the patentee contended that the skilled addressee would recognise that certain words that appeared in the relevant claim were included by mistake and that those words could be disregarded. As the Full Court observed at [111]-[112]:
[111] In the present case the primary judge found that the skilled addressee would understand that the reference to a reciprocating basket in claim 1 was an error and that he or she would have simply disregarded those words.
[112] GSK relies on the skilled addressee’s understanding of the claim to establish not what the language of the claim actually conveys to the skilled addressee, but for the purpose of ignoring some of the language used on the basis that it must have been included by mistake. The question is whether, if the court were to also interpret the claim so as to correct the mistake, it would be re-writing the claim or merely interpreting it through the eyes of the skilled addressee in accordance with the principles to which we have referred.
106 The Full Court upheld the primary judge’s decision which gave effect to the words which the patentee contended should be disregarded. I should make clear that ToolGen did not contend that the approach rejected by the primary judge and the Full Court in GlaxoSmithKline should be adopted in this case. I will return to consider the details of the construction issues shortly but it is important to observe that ToolGen’s case on the first issue of construction that arises for decision was that the language of claim 1 was capable of supporting the broad meaning which ToolGen sought to attribute to it. I have referred to GlaxoSmithKline not because the present case is said to involve any mistake in the drafting of claim 1, but merely to draw attention to the Full Court’s observations concerning the role, and limits, of purposive construction.
107 It is convenient to refer at this point to the Full Court decision in Novozymes A/S v Danisco A/S (2013) 99 IPR 417 (Greenwood, Jessup and Yates JJ) (“Novozymes”) which was relied on by ToolGen in its submissions as raising an issue of construction similar to one that arises here. The patent in suit in that case included 17 claims. Claims 1, 7 and 9 were for:
1. A process for preparing a foodstuff suitable for consumption comprising an emulsifier, the process comprising the steps of
(i) providing a food material containing a fatty acid ester and a second constituent;
(ii) contacting the food material with an enzyme such that an emulsifier is generated by the enzyme from the fatty acid ester and a second functional ingredient is generated from the second constituent;
(iii) inactivating or denaturing the enzyme to provide the foodstuff comprising the emulsifier, the fatty acid ester and the enzyme in an inactive form or a denatured form.
…
7. A process according to any one of the preceding claims wherein the foodstuff comprises at least the emulsifier and the second functional ingredient, and wherein the emulsifier and the second functional ingredient have been generated from the fatty acid ester and the second constituent of the food material by the enzyme.
…
9. A process according to any one of the preceding claims wherein the second constituent is selected from a constituent comprising a hydroxy group (-OH), polyvalent alcohols, including glycerol; water, ethanol, sugars including sucrose, fructose, glucose (dextrose), lactose, and galactose; dextrins including maltodextrin, sorbitol, mannitol, fruit acids and hydroxy acids including citric acid, tartaric acid, lactic acid and ascorbic acid; mixtures and derivatives thereof.
108 The primary judge in that case found that water could act as the “second constituent” in claim 1 but not claim 7. However, as Jessup J (with whom Yates J relevantly agreed) said at [70]:
[70] That conclusion turned upon the terms of claim 9, which is dependent on claim 1. Absent claim 9, the primary judge considered that water could not be the second constituent under claim 1. However, claim 9 specified water as within the range of permissible second constituents, and was consistent with the specification in doing so. It followed, according to her Honour, that the skilled addressee would understand that at least a permissible second constituent under claim 1 would be water. Her Honour gave instances, within the compass of the claim, in which water would be the second constituent…
109 It is not necessary to explore the intricacies of the claims and, in particular, the relationship between claims 1, 7 and 9. Importantly, however, Jessup J rejected the primary judge’s conclusion that the skilled addressee would read claim 7 as excluding a process in which there was one reaction only and water was the second constituent. His Honour continued at [87]-[88]:
[87] That brings me back to claim 9, the passage in the specification corresponding to which is set out in the third extract in [42] above. Under claim 9, what is claimed, among other things, is a process according to claim 7 in which water is permitted as a second constituent. If we are to read the claims sensibly, it would seem to follow that any ambiguity in claim 7 must be resolved by allowing water to be the second constituent. The primary judge deflected that conclusion by reasoning that, in the eye of the skilled reader, “the specificity of claim 7 would be understood to exclude water, even though it was one of the broad class of second constituents of claim 9”. The point was, it seems, that, although the wording of other claims might aid in the construction of the claim of interest, that was only an aid to construction, and had to yield to specific indications as to the meaning of the latter. In the context of the present debate, according to her Honour, it was clear from the words of claim 7 itself that water could not be the second constituent, and that conclusion was not to be displaced by the more indirect constructional indications that might be derived from another claim. A submission along these lines was also made by the respondents before the Full Court.
[88] It should be clear that I do not share her Honour’s perception as to the clarity of the specific meaning of claim 7 itself. But there is, in my respectful view, a more direct, and less tractable, impact which the terms of claim 9 have upon the construction of claim 7. Claim 9 bears upon the issue not merely because it provides a point of reference for the achievement of consistency of meaning within the patent as a printed document. More importantly, there is a structural relationship between claim 7 and claim 9: that the patent claims a process according to claim 7 in which water is the second constituent is the inevitable result of the way claim 9 is expressed. Claim 9 could not be clearer in relevant respects. It tells the reader, both scientific and lay, that water must be within the class of second constituents permissible under each of the preceding claims, including claim 7.
110 It is important to note that Jessup J did not consider that there was anything in claim 7 that expressly, or by necessary implication, excluded a process in which water was the second constituent. However, even if claim 9 was ambiguous as to whether or not it excluded water as the second constituent, the position was made clear by the presence of claim 9 which expressly contemplated that water could act as the second constituent.
111 His Honour went on to consider, against the background of those conclusions, the issue of clarity and, in particular, whether claims 1 and 7 were invalid because they were not “clear and succinct” as required by s 40(3) of the Act as it then stood. Neither the primary judge nor the Full Court considered claim 7 to be invalid for lack of clarity, although they disagreed as to how claim 7 was to be interpreted. On the issue of lack of clarity his Honour’s observations included the following at [93]:
[93] The problem of locating the dividing line which separates a claim which is bad for want of clarity from a claim which, though troublesome in its construction, is sufficiently clear to be valid, is scarcely less difficult than the problem of construction itself. In the present case, both sides have been able to draw upon general statements in the cases which provide some support for the opposite positions which they take. For my own part, I would find it sufficient for present purposes to refer to Welch Perrin & Co Pty Ltd v Worrel (1961) 106 CLR 588, in which the High Court said (at 610):
If it is impossible to ascertain what the invention is from a fair reading of the specification as a whole, that, of course, is an end of the matter. But this objection is not established by reading the specification in the abstract. It must be construed in the light of the common knowledge in the art before the priority date. The general principles governing the construction of specifications are well known, and no lengthy reference to them is necessary. It is, however, fitting that we remind ourselves of the criterion to be applied when it is said that a specification is ambiguous. For, as the Chief Justice pointed out in Martin v Scribal (1954) 92 CLR 17 at 59, referring to Lord Parker’s remarks in National Colour Kinematograph Co Ltd v Bioschemes Ltd (1915) 32 RPC 256, we are not construing a written instrument operating inter partes, but a public instrument which must, if it is to be valid, define a monopoly in such a way that it is not reasonably capable of being misunderstood. Nevertheless, it is to be remembered that any purely verbal or grammatical question that can be resolved according to ordinary rules for the construction of written documents, does not, once it has been resolved, leave uncertain the ambit of the monopoly claimed (see Kauzal v Lee (1936) 58 CLR 670 at 685).
112 I should note in relation to his Honour’s reference to Welch Perrin & Co Pty Ltd v Worrell (1961) 106 CLR 588 that the High Court is not to be understood as suggesting (as his Honour no doubt appreciated) that uncertainty of meaning can always be resolved by application of the rules for the construction: see Freeman v TJ and FL Pohlner Pty Ltd (1994) 30 IPR 377 at 381-382.
113 The first construction issue concerns the references to “nucleic acid encoding” in claims 1 and 10 and their dependent claims. Claims 1 and 10 both use the phrase “a nucleic acid encoding” when referring to “a nucleic acid encoding a Cas9 polypeptide” (component 1) and again when referring to “a nucleic acid encoding a guide RNA” (component 2). The debate between the parties was primarily concerned with the meaning of the phrase as used in component 2.
114 The respondents submitted that ToolGen’s construction of the word “encoding” when referring to a nucleic acid encoding a guide RNA in claims 1 and 10 involves giving the word “encoding” a meaning which is different from its ordinary use in the field of molecular biology. But ToolGen argues (correctly) that this does not in itself exclude the possibility that the word is used in claim 1 and 10 in a different sense. ToolGen says that the word encoding is also apt to mean, in the context in which it is used here, “providing the sequence for” a guide RNA. It says that the word has a broader meaning which includes not only a string of nucleotides that are transcribed into a guide RNA, but also a string of nucleotides that enables the guide RNA to perform its function (i.e. to guide a Cas9 protein). As such, ToolGen says no act of transcription within the cell to produce the guide RNA is required by the use of the word “encoding” and, further, if there must be an act of transcription, then it is not one that must occur inside the eukaryotic cell in which the target nucleic acid sequence resides. According to ToolGen, “nucleic acid encoding a guide RNA” in claims 1 and 10 includes both DNA which is transcribed to RNA in vivo in a eukaryotic cell and RNA which is transcribed in vitro prior to it being introduced into the eukaryotic cell and which provides the information for the guide RNA to perform its function of guiding the cas9 to the target DNA once inside a eukaryotic cell.
115 ToolGen placed considerable reliance on claim 19 which, when read with claim 10, requires that the nucleic acid encoding the guide RNA is in vitro transcribed RNA. According to ToolGen, claim 19 makes no sense if “encoding” refers to the process of transcription because a nucleic acid “encoding” (in the conventional sense of the word) the guide RNA could only be DNA whereas claim 19 requires that the nucleic acid encoding the guide RNA is in vitro transcribed RNA. On that view, claim 19 refers to a situation in which in vitro transcribed RNA transcribes the guide RNA. The experts agreed that does not make sense.
116 ToolGen says if the words “a nucleic acid encoding a guide RNA” are understood as a nucleic acid “providing the sequence for” a guide RNA, then claim 10 and claim 19 can be read together. On that approach, claim 19 requires that in vitro transcribed RNA provide the sequence for the guide RNA.
117 ToolGen also placed considerable reliance on the fact that nucleic acid may be either DNA or RNA. It submitted that the wider term used in the claims should be presumed to have been deliberately chosen. As I understand the argument, use of the term nucleic acid rather than DNA is consistent with the language of claim 19 which requires that the nucleic acid encoding the guide RNA is in vitro transcribed RNA. ToolGen says that since RNA cannot be transcribed into guide RNA, the word “encoding” as used in claims 1, 10 and 19, should not be given its conventional meaning.
118 As previously mentioned, the respondents submitted, and the Delegate found, that claim 19 lacks clarity. The respondents say that the phrase “a nucleic acid encoding” as used in claim 10 is conventional language that would be used by a molecular biologist to describe the processes of transcription in which the guide RNA is encoded by DNA. The respondents say that ToolGen’s argument based on claim 19 is one in which “the tail wags the dog”.
119 In support of its submissions, ToolGen relied on the Full Court’s decision is Novozymes to which I have previously referred. In my opinion, that case merely reflects the application of the ordinary principles of construction and, in particular, the need to read the complete specification, including the claims, as a whole. In the present case, the question is what claims 1, 10 and 19 mean in light of the complete specification when read as a whole through the eyes of the skilled addressee. The process of construction necessarily includes a consideration of claim 19 and the implications that its presence may have for claims 1 and 10.
120 There are two points to make about the relationship between claims 1, 10 and 19. First, claim 1 is for a composition and claim 10 is for a method. I consider the skilled addressee would understand that claim 1 defines a composition suitable for use in carrying out the method of claim 1. They are two aspects of the same invention. The second point is that claim 19 is dependent on claim 10, but not claim 1. It is the relationship between claim 10 and claim 19 that is at the core of ToolGen’s argument.
121 Claim 10 makes clear that the method of the claim requires that the nucleic acid encoding a polypeptide and the nucleic acid encoding a guide RNA do the encoding in the eukaryotic cell in which the target nucleic acid sequence is located. The word encoding is used as a transitive verb to describe the relationship between the relevant action (encoding), the subject (the nucleic acid) and the object (a polypeptide or a guide RNA). That the relevant action occurs inside the cell is made clear by the introductory words of claim 10 which refer to a method in which the CRISPR/Cas system is introduced into the eukaryotic cell. That system comprises the nucleic acid encoding a Cas9 polypeptide (component 1) and the nucleic acid encoding a guide RNA (component 2). In my opinion, claim 10 requires that nucleic acid be introduced into the eukaryotic cell where it will then encode a polypeptide and a guide RNA.
122 ToolGen’s construction of claim 10 requires only that the nucleic acid “provide the sequence for a guide RNA”. On ToolGen’s construction of claim 10 when read with claim 19, the transcription of the guide RNA can also occur in vitro (outside the eukaryotic cell in which the target sequence resides). ToolGen’s argument is that the verb “encoding” can mean both providing the sequence for producing the guide RNA (through the process of transcription from DNA to RNA) as well as providing the sequence that enables the guide RNA to perform its function. I do not accept that the later interpretation involves any action at all as the guide RNA once produced from DNA in vitro does not provide the information for its guiding function in any meaningful sense, and merely exists as an RNA capable of guiding Cas9 to the target sequence. The action that the word “encoding” describes is that of producing the RNA which, according to the express language of claim 10, must occur within the eukaryotic cell. Accordingly, it cannot be said that in vitro transcribed RNA which is produced outside of the cell and introduced into the cell in a “naked” or “isolated” form is consistent with what is described in claim 10.
123 Furthermore, if the nucleic acid referred to in claim 10 is isolated RNA transcribed in vitro (ie. before introduction to the cell) then what is described, on ToolGen’s construction, is isolated RNA providing the sequence for the guide RNA or, more specifically, a guide RNA (transcribed in vitro) providing the sequence for a guide RNA. That leads to the rather odd result in which the isolated RNA and the guide RNA are one and the same thing.
124 ToolGen also submitted that the term “nucleic acid encoding” used in claims 1 and 10 is never used in the body of the specification to refer only to DNA encoding a guide RNA. Further, ToolGen submitted that the words “nucleic acid encoding” are deliberately broader and different from the words that are used in the body of the specification, which refers to “DNA that encodes the guide RNA”. ToolGen submitted that the different language used shows that there is an important difference between “nucleic acid encoding the guide RNA” and “DNA that encodes the guide RNA” beyond the fact that the latter language refers only to DNA and not to all forms of nucleic acid. ToolGen also referred to [176]-[179] of the Specification which I have previously set out including, in particular, the statement in [176] that “[t]he guide RNA may be transferred into a cell or an organism in the form of RNA or DNA that encodes the guide RNA”. However, this statement is clearly distinguishing between two different approaches, the first in which the guide RNA is transferred into the cell “in the form of RNA” and the second in which DNA that encodes the guide RNA is transferred into the cell.
125 As to the use of the word “encodes” in [176] and [177] rather than “encoding” as used in the claims, there is in my view no material difference between the two. Both refer to nucleic acid that encodes a guide RNA. If anything, the language of the claims confirms that the relevant action (encoding of the guide RNA) is occurring inside the cell.
126 ToolGen also drew attention to the final sentence in [179] which states, in effect, that the introduction of DNA encoding a guide RNA is not excluded. That statement when read in the context of the balance of [179] indicates that this approach is less preferred to the introduction of an isolated (or naked) guide RNA. According to [179], the preferable approach is that in which “[t]he guide RNA is preferably transferred to a cell in the form of isolated RNA rather than in the form of plasmid comprising encoding sequence for a guide RNA”. Those words are referring to the use of a plasmid to introduce into the cell DNA encoding the guide RNA. The Specification refers at [328] to some of the potential problems associated with the use of plasmid DNA including unwanted immune responses in plants and animals.
127 The word “encoding” is used in the Specification to distinguish nucleic acid encoding a Cas protein from a Cas9 protein. For example, [156] distinguishes between a Cas component “in the form of a protein or in the form of a nucleic acid encoding Cas protein”. It is apparent that in the latter case the DNA will be transcribed to mRNA which in turn will be translated to a Cas protein. The Specification elsewhere refers to “Cas protein-encoding nucleic acid” which may be in the form of a vector such as a plasmid: see, for example, [166], [207], [217], [222] and [224]. The Specification states at [199]:
Using Cas protein rather than a nucleic acid encoding Cas protein to induce a targeted mutagenesis is advantageous because exogeneous DNA is not introduced into an organism. Thus, the composition comprising Cas protein and a guide RNA may be used to develop therapeutics or value-added crops, livestock, poultry, fish, pets, etc.
128 However, while the first of these methods is said to have advantages, the Specification distinguishes repeatedly between use of a Cas protein and use of nucleic acid encoding a Cas protein: see, for example, [203], [207] and [215]. The Specification states at [217]:
In the present invention, a Cas protein-encoding nucleic acid or Cas protein and a guide RNA or DNA that encodes the guide RNA may be transferred into a cell by various methods known in the art …
Here again the Specification is distinguishing between a Cas protein and nucleic acid encoding for a Cas protein either of which may be transferred into the cell. The same distinction is drawn with respect to the guide RNA, which may be introduced into the cell in its naked or isolated form, or in the form of DNA that encodes the guide RNA. The Specification also refers to “guide RNA encoding plasmid”: see, for example, [319] and [440]. The latter paragraph distinguishes between the use of (inter alia) “synthetic guide RNA” (ie. naked or isolated RNA) and “guide RNA encoding plasmids”.
129 ToolGen submitted that it would be surprising if the patent applicant sought to protect the less preferred method while not seeking protection for the preferred method which uses isolated RNA transcribed in vitro.
130 In my opinion, ToolGen’s submission is answered by Lord Hoffmann’s speech in Kirin-Amgen and the other authorities referred to in Multigate which emphasise that a patentee may have a good reason for introducing a seemingly inexplicable limitation. Here, the language of claim 10 when read in the context of the Specification as a whole indicates that (for whatever reason) the claim has been limited to a method in which the nucleic acid encodes the guide RNA in the eukaryotic cell. In my opinion, this requires that the guide RNA be transcribed from nucleic acid in the eukaryotic cell.
131 At a more general level, and focusing on the relationship between claim 10 and 19, ToolGen submitted that the respondents’ interpretation of claim 19 does not make any sufficient attempt to read the two claims together so as to give them a clear and consistent meaning which also avoids the conclusion that claim 19 lacks clarity. ToolGen submitted, in effect, that claim 10 must be interpreted so as to accommodate the language of claim 19 in order to ensure that claim 19 is given meaning and work to do.
132 As to ToolGen’s reliance on the use of the term “nucleic acid” rather than DNA in claim 10, there are two matters which would explain why the broader term is used. The first concerns the transcription and translation of the Cas9 polypeptide which will involve both DNA and RNA (in particular mRNA) in the transcription and translation process. The second relates to the use of viral RNA. The experts agreed that the use of the term “nucleic acid” will cover a situation in which viral RNA is introduced into the cell which then encodes DNA which in turn encodes the guide RNA. Although the use of viral RNA is not discussed in the Specification, this method of producing RNA in the cell was common general knowledge at the priority date. For that reason I do not think the use of the term “nucleic acid” (rather than DNA) in claim 10 assists ToolGen’s argument.
133 The question is whether the presence of claim 19 justifies the conclusion that the words “a nucleic acid encoding a guide RNA”, when read in the context of the Specification as a whole, are reasonably capable of meaning simply “providing the sequence for” a guide RNA. ToolGen relied on the evidence of Associate Professors Firestein and Herold in support of this construction of the word “encoding” in claims 1 and 10.
134 In JER 1 Associate Professor Firestein said that he understood the relevant phrase to:
… encompass also the guide RNA itself, agnostic to the method by which the guide RNA was generated (eg. including when generated in the in vitro transcribed form).
135 In support of this understanding he referred to claim 18 in which the nucleic acid encoding the guide RNA is a vector and claim 19 in which the nucleic acid encoding the guide RNA is in vitro transcribed RNA.
136 In Firestein 1, Associate Professor Firestein said that he understood the relevant phrase to refer to the nucleic acid sequence or code that comprises the DNA itself or the DNA from which it can be expressed. He went on to say that the phrase is referring to the information (ie. code) provided by the nucleic acid sequence that defines the composition of the guide RNA.
137 Associate Professor Firestein was cross-examined at some length on this topic. His answers were quite often not directly responsive to the question asked. He referred to an interpretation of claims 1 and 10 in which nucleic acid encoding a guide RNA might be understood as encompassing RNA providing the sequence for the guide RNA. Of the phrase “nucleic acid encoding a guide RNA” Associate Professor Firestein said:
… the phrase doesn’t say nucleic acid transcribing a guide RNA, it says encoding a guide RNA, so I view the word encoding simply as providing the sequence for providing the code. And that may come through transcription, or it may come from the sequence itself.
138 Associate Professor Firestein agreed that this would not be typical usage in the field of molecular biology. But it is clear that he was conscious of the difficulty with this construction because, as a molecular biologist, he understands that synthetic RNA (ie. isolated or naked RNA) introduced into the cell does not encode the guide RNA. Moreover, on his interpretation of the relevant claims, the nucleic acid encoding the guide RNA and the guide RNA are precisely the same thing: ie. a sequence of non-coding RNA. In cross-examination Associate Professor Firestein eventually accepted that the use of the word encoding as meaning no more than providing a sequence or code was different from its ordinary meaning in molecular biology. But he went on to add that “encoding” can have a broader meaning outside the field of molecular biology.
139 At least in his oral evidence Associate Professor Firestein referred to this interpretation as a possible interpretation of the claims. In this regard, he gave the following evidence:
MR DIMITRIADIS: You referred to the use of encoding in a molecular biology context and in other contexts. May we take it from what you’ve said, Professor Firestein, that your broader interpretation of this claim involves giving a meaning to encoding which differs from its meaning in the context of molecular biology.
ASSOC PROF FIRESTEIN: Yes, I – well, as I’ve said before, it’s not the typical language that I would use, but at the same time, I wouldn’t want to speculate exactly what the authors meant when they used this phrase, because I do understand that there’s other ways to interpret it.
140 Moving to Associate Professor Herold’s evidence on this topic, he stated in Herold 1:
150. Upon reading the phrase “nucleic acid encoding a guide RNA” in claim 1, my immediate understanding is that the “nucleic acid” referred to is DNA, in the form of a vector or plasmid. DNA “encodes” the guide RNA because it is transcribed by the cell's machinery into the guide RNA.
151. I would not typically consider the “nucleic acid encoding a guide RNA” to refer to the guide RNA itself. This is because it would not be my usual use of “encoding” to speak of “RNA encoding a guide RNA”.
152. However, I note that claim 19 of the Patent Application is directed to the “method of any one of claims 10-16, wherein the nucleic acid encoding the guide RNA is in vitro transcribed RNA”. If claim 19 is within the scope of claim 10, then this indicates that the phrase “nucleic acid encoding a guide RNA”, at least in claim 10, is intended to include RNA itself. I expect the words to have the same meaning in claim 1. Claims 9, 20 and 21, which I discuss below, also appear to be consistent with the phrase “nucleic acid encoding a guide RNA” in claims 1 and 10 including in vitro transcribed RNA.
153. As stated above, nearly all of the examples are directed to in vitro transcribed guide RNA, and the experimental detail in relation to DNA encoding the guide RNA is very limited. Although DNA (plasmids) encoding chimeric sgRNA are referred to in Examples 1-3 and 6, there are no details about how the plasmids are made.
141 He restated this view in JER 1 by referring to those same paragraphs of Herold 1. In his oral evidence he agreed that claim 19 makes sense if the broader construction of the word “encoding” is adopted. But he was clear that this involves giving the word a different meaning to that which it is given in the field of molecular biology. In particular, he agreed with the following evidence given by Professor Thomas in concurrent evidence:
MR DIMITRIADIS: … Professor Thomas, you understand that Professor Firestein’s broader interpretation of the claim is that it – in the phrase “nucleic acid encoding a guide RNA encompasses the guide RNA itself, such as a in vitro transcribed guide RNA”. Is it your view not that that interpretation of the claim involves giving the word “encoding” its ordinary meaning in molecular biology as at October 2012, or some other meaning?
PROF THOMAS: I think Professor Firestein’s view involved attaching a meaning to “encoded” [sic] that is not one that is used in molecular biology. And further, I would say that given that the phrase starts “nucleic acid”, we are definitely in the realms of molecular biology when looking at this particular phrase.
MR DIMITRIADIS: Professor Herold, can I ask you do you agree or not with Professor Thomas’ evidence on that point?
PROF HEROLD: Yes. I do agree.
142 My view of Associate Professor Herold’s evidence is that he has approached the interpretation of claim 10 on the basis that claims 10 and 19 must be read consistently and that this is only possible if the words “nucleic acid encoding a guide RNA” encompass the introduction of isolated or naked RNA into the cell. In my opinion his approach overlooks the possibility that claim 19 cannot be sensibly read with claim 10 and that the meaning of claim 19 when read with claim 10 is unclear. Of the molecular biologists who gave evidence on this topic, I prefer the evidence of Professor Thomas. In my opinion, his evidence is likely to be representative of the notional skilled addressee who, when reading the Specification as a whole in light of the common general knowledge, would have a very clear understanding of the system described in claim 10 as one in which nucleic acid encoding the guide RNA is introduced into the cell where the guide RNA is transcribed. As Professor Thomas said in his oral evidence in relation to the words nucleic acid encoding a guide RNA:
PROF THOMAS: … for me, the statement “nucleic acid encoding a guide RNA” does not encompass the guide RNA itself. To me, that’s a very clear molecular biology statement in which we have a DNA or RNA sequence that provides the information for the guide RNA. So I would be stronger in my statement.
What I understood Professor Thomas to be saying is that the nucleic acid provides the information from which the guide RNA is transcribed. Claim 19 does not make sense to Professor Thomas because he does not consider that in vitro transcribed guide RNA is a nucleic acid that encodes anything. I regard his evidence on this topic as highly persuasive.
143 The relevant paragraphs of the body of the Specification, including the description of a system in which nucleic acid (DNA) encodes the guide RNA in the eukaryotic cell, and the language used to describe that system, weigh heavily against ToolGen’s construction. In my opinion, the word “encoding” is used in claim 10 in its conventional sense (ie. as it would be understood by a molecular biologist) to refer to the production of a Cas9 polypeptide by transcription and translation and the production of a guide RNA by transcription in the cell. The nucleic acid referred to in the claim provides the information which is used in the cell to produce the guide RNA. Claim 10 does not encompass a system in which an existing guide RNA is introduced into the cell.
144 As Jessup J observed in Novozymes at [95]:
[A] conclusion that a claim lacks clarity is proper to be made only if the court, using all the properly available aids and looking at the matter through the eyes of the skilled addressee, is unable to give a clear meaning to the claim.
145 Claim 10 requires that the guide RNA be transcribed in the cell but claim 19 contemplates that the guide RNA will have been transcribed outside the cell. Claim 19 cannot be read sensibly with claim 10 on which it is dependent. In my opinion claim 19 lacks clarity.
146 I have previously referred to the definition of “paired Cas9 nickase” in the Specification which refers to the guide RNA and Cas9 protein functioning as a pair. The Specification states at [187]:
The guide RNA and the Cas protein may function as a pair. As used herein, the term “paired Cas nickase” may refer to the guide RNA and the Cas protein functioning as a pair. The pair comprises two guide RNAs. The guide RNA and Cas protein may function as a pair, and induce two nicks on different DNA strand. The two nicks may be separated by at least 100 bps, but are not limited thereto.
147 The definition is somewhat awkward but the experts were in agreement as to what a paired Cas9 nickase is in terms broadly consistent with the definition. A composition that uses paired Cas9 nickases is one in which there are two different guide RNAs each guiding a Cas9 nickase. Each nickase produces a single break in a strand of DNA. A pair of nickases produces two such breaks.
148 The question is whether either claim 1 or claim 10 encompasses paired Cas9 nickases. Although the Specification refers to paired Cas9 nickases, including in the definition to which I have referred and in Examples 7 and 8, none of the claims made any explicit reference to them. There are two main points of contention in relation to this construction issue. The first is whether a composition that includes paired Cas9 nickases that produce staggered ended breaks at two different positions on two DNA strands is capable of being said to produce a “site-specific double-stranded break” as required by claims 1 and 10. The second is whether a composition that includes paired Cas9 nickases which uses two different RNAs paired with two Cas9 nickases can be correctly characterised as “a Cas9/RNA complex” as required by claim 10.
149 To give some context to this discussion, I should note that the respondents contend that claim 1 extends to such a composition, but that the disclosure of P1 does not. It follows according to the respondents’ argument that claim 1 is not entitled to priority based on P1. The same argument is relied on by the respondents in relation to the method defined by claim 10.
150 Professor Thomas considered that both claim 1 and claim 10 use language that extends to the use of paired Cas9 nickases. He did not draw any relevant distinction between claim 1 and claim 10 in this respect. While Professor Thomas accepted that the language used to denote both the guide RNA and the Cas9 polypeptide are expressed in the singular, he understood both claim 1 and claim 10 to not exclude the use of two different complexes consisting of two different guide RNAs and two Cas9 nickases to generate a double-stranded break.
151 Senior Counsel for ToolGen put to Professor Thomas that the claims referred to the introduction of a site-specific double-stranded break, and that this was inconsistent with the use of paired Cas9 nickases that would produce breaks at different nucleotides on each strand. He did not accept this. In his view, the nickase system produces a site-specific double-stranded break at different positions on the two DNA strands. Associate Professor Herold agreed that a site-specific double-stranded break could be either blunt (meaning that the breaks were at the same location on each DNA strand) or staggered (meaning that the breaks could occur at different locations).
152 ToolGen submitted that the concept of “site-specific” in relation to a DNA strand means a particular nucleotide position on that strand. ToolGen submitted that if the breaks occur at two different nucleotide positions on the two strands, then the break in the DNA is not “a site-specific double-stranded break” but “a two site-specific double-stranded break” outside the scope of the claims. I do not accept that submission which, in my opinion, relies on an overly literal analysis of the claim language that is inconsistent with what I consider would be the notional skilled addressee’s understanding of this requirement.
153 Professor Thomas said that the nickase system would produce a site-specific double-stranded break where the breaks on each strand are in close proximity. He said that if the breaks are not in close proximity there will be large overhangs which will prevent the two strands from separating (hence the name “sticky ends”). He gave the following evidence:
PROF THOMAS: Yes, I believe, based on my knowledge of the way that the nickase system works, that the nicks must be in relatively close proximity to generate a double stranded break, and that they’re separated by a distance, in this case the reference to 100 based pairs or more, then that will not constitute a double – an effective – double stranded break, because the strands don’t separate.
…
MR CORDINER: … Would you agree with me … a more natural understanding of a paired cas9 nickases, is that it doesn’t produce a site specific double stranded break. It produces a double stranded break, but not a site-specific one.
PROF THOMAS: I can’t agree with you there, actually. I wouldn’t say – so in terms of a site-specific double stranded break, each of the nicks that occur is occurring at a site-specific location, and if they’re in close proximity, they would generate a double stranded break. So to me, the nickase system does include a site-specific double stranded break.
MR CORDINER: Yes. But I put it to you that another way of interpreting that, given what the specification has said, it’s not unreasonable to hold the view that the reference to … a site-specific is really referring to the fact that this is happening at a single nucleotide.
PROF THOMAS: No, again, I can’t agree with you there, in the sense that it is a single – it is a very specific location that is targeted by each of the nickases, so it’s a – it’s – the term site-specific totally encompasses, in this – in this context, to me, a double stranded break generated by two nickases on each strand of the DNA in close proximity.
Associate Professor Herold was also of the view that a site-specific double-stranded break could be one with staggered end breaks.
154 In his evidence, Professor Thomas made clear that the breaks would need to be closely proximate to each other. ToolGen says that the difficulty with this approach is that there is no requirement in the claims that the two breaks be any particular distance from each other. However, Professor Thomas’ evidence was that if the breaks are too far apart, there will be no effective double-stranded break because the two strands will not separate.
155 Associate Professor Firestein, who did not consider either claims 1 or 10 to extend to paired Cas9 nickases, saw some difficulty with the language of “site-specific double-stranded break” but he agreed that this was not the reason behind his view. He explained the difficulty with that phrase and the reason he considered paired Cas9 nickases were outside the claims in the following evidence:
MR DIMITRIADIS: Professor Firestein, just to back up that point, the use of the phrase “site-specific” in claims 1 and 10 is not the reason why you regard the use of paired Cas9 nickases as being outside the claim, it’s for a different reason that you regard them as being outside the claim. Correct?
ASSOC PROF FIRESTEIN: That’s correct.
MR DIMITRIADIS: And you would agree, would you not, that the action of a paired Cas9 nickase is site-specific in the sense that that system will lead to the introduction of a double-stranded break at a particular location in the DNA sequence. Correct?
ASSOC PROF FIRESTEIN: I would not necessarily agree with that because – you know, I think the definition of what is site-specific is – can be a bit nebulous and confounding. As the patent application itself shows and discusses, the distance between the two components on the paired nickase system can be quite close. They could be 100 base pairs, they could be 1000 base pairs, they can be 10,000 base pairs apart. So at what point does it become site-specific? If you have two guide RNA Cas9 complexes that are potentially 10,000 base pairs apart, is that site-specific, or are we talking about two sites? So, to me, the definition of site-specific would be a bit confusing, so I interpret more strongly the use of the words around the complex, being a complex and a guide RNA rather than the interpretation around the term “site-specific.”
MR DIMITRIADIS: Yes. It’s the latter point that is your reason for excluding the Cas9 nickases from the claims. Correct?
ASSOC PROF FIRESTEIN: Yes. It’s really about the fact that you have two different heterologous complexes that are working in the paired system.
156 Associate Professor Herold considered that claim 1 encompassed a paired Cas9 nickase but that claim 10 did not. With regard to claim 10, he appears to have given particular weight to the last integer of that claim and the requirement that “the Cas9 polypeptide and guide RNA [which] form a Cas9/complex in the eukaryotic cell, whereby a site-specific double-stranded break at the target nucleic acid sequence is introduced”. This language is different from the language of claim 1.
157 I do not think the term “site-specific” requires that the double-stranded break occur at a singular nucleotide position on both strands. All experts accepted that the term extended to staggered ended breaks occurring at different nucleotide positions on the two DNA strands. Both Professor Thomas and Associate Professor Herold were of this view. Associate Professor Firestein had some difficulty with the meaning of site-specific, but he did not suggest that a site-specific (see his oral evidence quoted above) double-stranded break must occur at the same nucleotide position on each strand. Accordingly, I find that the language of claims 1 and 10 does extend to staggered ended breaks that are in close proximity.
158 However, I do not think that either claim 1 or claim 10 encompass paired Cas9 nickases for the reasons identified by Associate Professor Firestein. This is another situation in which the patent applicant has claimed less than what it disclosed in the body of the Specification. The use of the singular to define the Cas9 polypeptide and the guide RNA coupled with a proper understanding of the role each of the guide RNA and the polypeptide play is consistent with a description of a complete system in which a single guide RNA and a single Cas9 polypeptide combine to introduce a site-specific double-stranded break in DNA. Claim 1 does not extend to what I consider is a different system utilising a pair of Cas9 nickases each with its own guide RNA. The language of claim 10 makes very clear that what is being described in claim 10 is a complex consisting of a guide RNA and a Cas9 polypeptide which create a double-stranded break. In this respect, claim 1 and claim 10 are describing the same invention but in slightly different language that, apart from the fact that one is for a composition and the other a method, are not relevantly different.
159 The respondents placed some reliance in their written submissions on the definition of the words “comprising” and “comprises” which appear at page 61 of the Specification. It stated that, unless the context requires otherwise, those words do not imply the exclusion of any other integer or group of integers. That definition does not assist the respondents for two reasons. First, as the respondents would have me read them, the claims include a system for introducing a site-specific double-stranded break that is different from the system that is described, and not merely by reason of the addition of further integers. On the respondents’ construction of the claims, each pair of guide RNAs and nickases produces a single rather than a double break. The function performed by each nickase is in that respect materially different from the function performed by a single guide RNA and endonuclease. Giving effect to the definition in those circumstances would have implications for the validity of the claim. Secondly, and relatedly, in my view “the context requires otherwise” and, therefore, the definition does not apply.
RELEVANT LEGISLATIVE PROVISIONS
160 Section 40(2) and (3) of the Act relevantly provide:
Requirements relating to complete specifications
(2) A complete specification must:
(a) disclose the invention in a manner which is clear enough and complete enough for the invention to be performed by a person skilled in the relevant art; and
…
(3) The claim or claims must be clear and succinct and supported by matter disclosed in the specification.
161 Section 43(2) and (2A) of the Act provide:
(2) The priority date of a claim is:
(a) if subsection (2A) applies to the claim—the date determined under the regulations; or
(b) otherwise—the date of the filing of the specification.
(2A) This subsection applies to a claim if:
(a) prescribed circumstances apply in relation to the invention defined in the claim; and
(b) a prescribed document discloses, or a prescribed set of prescribed documents considered together disclose, the invention in the claim in a manner that is clear enough and complete enough for the invention to be performed by a person skilled in the relevant art.
162 Regulation 3.13A(2) of the Patents Regulations 1992 (Cth) relevantly provides that the “prescribed circumstances” are where a PCT application claims priority of an earlier basic application made no more than 12 months before the filing date of the PCT application. It is common ground that P1 is such an application. Given the patent application is a PCT application, reg 3.13A applies. Regulation 3.13A(1)(b)(i) requires that the earlier basic application (“the prescribed document”) be a document that “clearly discloses the invention in the claim”.
163 Regulation 3.12(4) provides that in “this Division [which includes reg 3.13A], a document … clearly discloses an invention if the document, or set of documents, discloses the invention in a manner that is clear enough, and complete enough, for the invention to be performed by a person skilled in the relevant art”.
164 Accordingly, reg 3.12(4) deems that reg 3.13A(1)(b)(i) will be satisfied where the earlier basic application “discloses the invention in a manner that is clear enough, and complete enough, for the invention to be performed by a person skilled in the relevant art” (“the priority date test”).
165 There is no additional requirement imposed by the words “clearly discloses” in reg 3.13A(1)(b)(i). Notably, the priority date test is the same as that set out not only in s 43(2A)(b) of the Act but also s 40(2)(a) (“the sufficiency test”). There is no longer a requirement for the claimed invention to be fairly based on matter disclosed in the priority document.
166 Section (40)(2)(a) requires that the complete specification make the necessary disclosure. Since the claims form part of the complete specification they may contribute to the disclosure. However, for the purposes of s 43(2A), the claims in the complete specification, although defining the invention, do not contribute to the disclosure. They serve only to define the invention which must be disclosed in the priority document “… in a manner that is clear enough and complete for the invention to be performed by a person skilled in the relevant are”. That said, the language used in s 40(2)(a) and s 43(2A)(b) is essentially the same: in both cases the invention must be disclosed in a manner that is clear enough and complete enough for the invention to be performed by a person skilled in the relevant art. Section 43(2A)(b) expresses the disclosure obligation by reference to the invention “in the claim”. In my view, nothing turns on the absence of the words “in the claims” in s 40(2)(a). The disclosure obligation imposed by s 40(2)(a) of the Act relates to the invention as claimed.
167 The Explanatory Memorandum to the RTB Act made it clear that the new s 40(2)(a) of the Act:
… is intended to align the disclosure requirement with that applying in other jurisdictions with the effect that sufficient information must be provided to enable the whole width of the claimed invention to be performed by the skilled person without undue burden, or the need for further invention …
An alternative to the existing Australian description requirement is the more stringent requirement that the skilled person reading the specification must be able to perform the invention across the whole width of the claims, not merely in relation to one among other embodiments within their scope. This requirement is consistent with the principle that the description accords with the scope of the monopoly granted.
The item is intended to modify the wording of paragraph 40(2)(a) of the Act so as to require enablement across the full width of the claims, while adopting language that is consistent with that used in other jurisdictions. The wording in the amendment is similar to s 14(3) of the UK patents legislation … The wording is also similar to art 83 of the European Patent Convention which has been interpreted with similar effect. The intention is that paragraph 40(2)(a) be given, as close as is practicable, the same effect as the corresponding provisions of UK legislation and the European Patent Convention.
(Emphasis added. Footnotes omitted.)
168 It is apparent that the amendments to s 40(2)(a) of the Act were made with the intention of aligning the Australian law of sufficiency with UK and European law. In particular, the old law, which had generally been held to require the enablement of only a single embodiment of the invention within each claim, was done away with.
169 Article 83 of the European Patent Convention (“EPC”) provides:
The European patent application shall disclose the invention in a manner sufficiently clear and complete for it to be carried out by a person skilled in the art.
170 Article 100(b) of the EPC provides that it is a ground of opposition that:
the European patent does not disclose the invention in a manner sufficiently clear and complete for it to be carried out by a person skilled in the art;
171 Sections 14(3) and 72(1)(c) of the Patents Act 1977 (UK) (“the UK Act”) correspond to Articles 83 and 100(b) of the EPC respectively.
172 In Eli Lilly & Co v Human Genome Sciences Inc [2008] RPC 29 (“Eli Lilly”) Kitchin J (as his Lordship then was) referred to the key elements of the sufficiency requirement at [239]-[243]:
[239] The specification must disclose the invention clearly and completely enough for it to be performed by a person skilled in the art. The key elements of this requirement which bear on the present case are these:
(i) the first step is to identify the invention and that is to be done by reading and construing the claims;
(ii) in the case of a product claim that means making or otherwise obtaining the product;
(iii) in the case of a process claim, it means working the process;
(iv) sufficiency of the disclosure must be assessed on the basis of the specification as a whole including the description and the claims;
(v) the disclosure is aimed at the skilled person who may use his common general knowledge to supplement the information contained in the specification;
(vi) the specification must be sufficient to allow the invention to be performed over the whole scope of the claim;
(vii) the specification must be sufficient to allow the invention to be so performed without undue burden.
[240] Elements (vi) and (vii) merit a little elaboration. It has long been a principle of patent law that the specification must enable the invention to be performed to the full extent of the monopoly claimed. If the invention discloses a principle of general application, the claims may be in correspondingly general terms. But if the claims include a number of discrete methods or products, the patentee must enable the invention to be performed in respect of each of them: Genentech I/Polypeptide expression T 292/85 [1989] OJEPO 275; Biogen Inc v Medeva plc [1997] R.P.C. 1 at [48].
[241] The question whether a burden is undue must be sensitive to the nature of the invention, the abilities of the skilled person and the art in which the invention has been made. The court must consider whether the effort required of the skilled person is undue having regard to the fact that the specification should explain to him how the invention can be performed. Each case must be decided on its own facts. Nevertheless, helpful guidance is to be found in the decision of the Court of Appeal in Mentor Corp. v Hollister Inc [1993] R.P.C. 7. Lloyd L.J. explained at 13–14:
“ …. if a working definition is required then one cannot do better than that proposed by Buckley L.J. in giving the judgment of the Court of Appeal in Valensi v British Radio Corporation [1973] R.P.C. 337. After referring to a number of earlier authorities, including Edison & Swan v Holland, he said:
‘We think that the effect of these cases as a whole is to show that the hypothetical addressee is not a person of exceptional skill and knowledge, that he is not to be expected to exercise any invention nor any prolonged research, inquiry or experiment. He must, however, be prepared to display a reasonable degree of skill and common knowledge of the art in making trials and to correct obvious errors in the specification if a means of correcting them can readily be found.’
Then a little later:
‘Further, we are of the opinion that it is not only inventive steps that cannot be required of the addressee. While the addressee must be taken as a person with a will to make the instructions work, he is not to be called upon to make a prolonged study of matters which present some initial difficulty: and, in particular, if there are actual errors in the specification—if the apparatus really will not work without departing from what is described—then, unless both the existence of the error and the way to correct it can quickly be discovered by an addressee of the degree of skill and knowledge which we envisage, the description is insufficient.’
In that case there was a mistake in the specification. But Buckley L.J.’s language is equally apt to cover an omission.
….
Before leaving the authorities, I should mention No-Fume Ltd v Frank Pitchford & Co Ltd (1935) 52 R.P.C. 231. Quoting from the judgment of Romer L.J. in that case, Buckley L.J. in Valensi is reported as saying:
‘The test to be applied for the purpose of ascertaining whether a man skilled in the art can readily correct the mistakes or readily supply the omissions, has been stated to be this: Can he rectify the mistakes and supply the omissions with the exercise of any inventive faculty? If he can, then the description of the specification is sufficient. If he cannot, the patent will be void for insufficiency.’
‘With’ in that quotation must be a misprint for ‘without’.
I turn to the judge’s conclusion on the law. What he said was this:
‘The section requires the skilled man to be able to perform the invention, but does not lay down the limits as to the time and energy that the skilled man must spend seeking to perform the invention before it is insufficient. Clearly there must be a limit. The sub-section, by using the words, clearly enough and completely enough, contemplates that patent specifications need not set out every detail necessary for performance, but can leave the skilled man to use his skill to perform the invention. In so doing he must seek success. He should not be required to carry out any prolonged research, enquiry or experiment. He may need to carry out the ordinary methods of trial and error, which involve no inventive step and generally are necessary in applying the particular discovery to produce a practical result. In each case, it is a question of fact, depending on the nature of the invention, as to whether the steps needed to perform the invention are ordinary steps of trial and error which a skilled man would realise would be necessary and normal to produce a practical result.’
I have already quoted the remainder of that paragraph.
I can find no vestige of error in that statement of the law. It was at first argued that the skilled man should not have to carry out any research, enquiry or experiment at all, whether prolonged or otherwise. But Mr. Thorley subsequently retreated from that extreme position. There is no support for setting so high a standard of disclosure, whether in Valensi itself or in any of the previous authorities, save possibly the judgment of Lindley L.J. in Edison & Swan v Holland. When, a little later, Aldous J. came to apply the law to the facts of this case, he refers to ‘routine trials’ and ‘normal routine matters that the skilled man would seek to do and would be able to do’. Mr. Thorley criticises the use of the word ‘routine’. To require the performance of routine trials is, he said, to ask too much of the addressee. I do not agree. ‘Routine’ is just the word I would have chosen myself to describe the sort of trial and error which has always been regarded as acceptable; and ‘routine trials’ has the further advantage that it is a positive concept, which is easily understood and applied. In practice, therefore, it may provide a surer test of what is meant by ‘clearly enough and completely enough’ in s.72(1) of the Act than the negative test proposed in Valensi, namely the absence of prolonged research, enquiry and experiment. If the trials are unusually arduous or prolonged, they would hardly be described as routine.”
[242] A little later (at page 17, lines 4–14) the court accepted the requirement was not to produce a successful commercial product but rather a workable prototype. This is an important point and one which must be kept well in mind in assessing inventions in the pharmaceutical field as much as any other.
[243] The case law of the EPO is, I believe, entirely consistent. Even though a reasonable amount of trial and error is permissible, when it comes to sufficiency of disclosure, for example in an unexplored field or where there are many technical difficulties, the skilled person has to have at his disposal, either in the specification or on the basis of his common general knowledge, adequate information leading necessarily and directly towards success through the evaluation of initial failures: see eg Unilever (1987) T 226/85.
173 Article 83 of the EPC was considered by the United Kingdom Supreme Court in Regeneron Pharmaceuticals Inc v Kymab Ltd [2020] UKSC 27 (“Regeneron”). Lord Briggs (with whom Lord Reed, Lord Hodge and Lord Sales agreed) said at [56]:
[56] Reflection upon those European and UK authorities yields the following principles:
(i) The requirement of sufficiency imposed by article 83 of the EPC exists to ensure that the extent of the monopoly conferred by the patent corresponds with the extent of the contribution which it makes to the art.
(ii) In the case of a product claim, the contribution to the art is the ability of the skilled person to make the product itself, rather than (if different) the invention.
(iii) Patentees are free to choose how widely to frame the range of products for which they claim protection. But they need to ensure that they make no broader claim than is enabled by their disclosure.
(iv) The disclosure required of the patentee is such as will, coupled with the common general knowledge existing as at the priority date, be sufficient to enable the skilled person to make substantially all the types or embodiments of products within the scope of the claim. That is what, in the context of a product claim, enablement means.
(v) A claim which seeks to protect products which cannot be made by the skilled person using the disclosure in the patent will, subject to de minimis or wholly irrelevant exceptions, be bound to exceed the contribution to the art made by the patent, measured as it must be at the priority date.
(vi) This does not mean that the patentee has to demonstrate in the disclosure that every embodiment within the scope of the claim has been tried, tested and proved to have been enabled to be made. Patentees may rely, if they can, upon a principle of general application if it would appear reasonably likely to enable the whole range of products within the scope of the claim to be made. But they take the risk, if challenged, that the supposed general principle will be proved at trial not in fact to enable a significant, relevant, part of the claimed range to be made, as at the priority date.
(vii) Nor will a claim which in substance passes the sufficiency test be defeated by dividing the product claim into a range denominated by some wholly irrelevant factor … The requirement to show enablement across the whole scope of the claim applies only across a relevant range. Put broadly, the range will be relevant if it is denominated by reference to a variable which significantly affects the value or utility of the product in achieving the purpose for which it is to be made.
(viii) Enablement across the scope of a product claim is not established merely by showing that all products within the relevant range will, if and when they can be made, deliver the same general benefit intended to be generated by the invention, regardless how valuable and ground-breaking that invention may prove to be.
174 Both Eli Lilly and Regeneron were revocation actions. The question was therefore whether the specification as a whole made a disclosure sufficient to enable the notional skilled addressee to perform the invention (leaving aside de minimis or irrelevant exceptions) across the scope of the relevant claims. What the invention is depends on the proper construction of the claim. Once the claim is construed it is then necessary to determine whether performance of the invention as claimed is enabled.
175 In the case of the patent application the same question must be addressed. The claims must be construed for the purpose of identifying the invention. It is then necessary to consider whether performance of the invention by the person skilled in the relevant art is sufficiently enabled by the complete specification. Although s 40(2)(a) of the Act does not expressly refer to the invention as the invention defined by the claim, or the invention in the claim (cf. s 43(2A)(b) of the Act), there is no reason to doubt that the sufficiency of the disclosure is to be assessed against the invention as claimed. This is confirmed by those parts of the Explanatory Memorandum to which I have referred which describe the requirement that the skilled person reading the specification be able to perform the invention across the whole width of the claims.
176 On the topic of priority dates and the requirements of s 43(2A) of the Act, the Explanatory Memorandum to the RTB Act stated:
Applicants should not be able to secure a priority date on the basis of a disclosure in a provisional or other relevant application that is less complete than required in a complete specification. Otherwise the applicant is in a position to deter competitors before they have fully realised the invention. Since an enabling disclosure will be required, the amendment will also align the requirements for securing a priority date with most other major patent jurisdictions.
177 Article 87 of the EPC is concerned with priority right. It provides:
Any person who has duly filed, in or for
(a) any State party to the Paris Convention for the Protection of Industrial Property or
(b) any Member of the World Trade Organization,
an application for a patent, a utility model or a utility certificate, or his successor in title, shall enjoy, for the purpose of filing a European patent application in respect of the same invention, a right of priority during a period of twelve months from the date of filing of the first application.
178 The right of priority referred to is available only where the first application (ie. the priority document) is “in respect of the same invention” as the European patent application. That requirement was considered by the English Court of Appeal in MedImmune Ltd v Novartis Pharmaceuticals UK Ltd [2013] RPC 27 (“MedImmune”). Kitchin LJ (with whom Moore-Bick and Lewison LJJ agreed) referred to the relevant principles as follows at [152]-[154]:
[152] The requirement that the earlier application must be in respect of the same invention was explained by the enlarged Board of Appeal of the EPO in G02/98, X/Same Invention, [2001] OJ EPO 413, [2002] E.P.O.R. 17:
‘The requirement for claiming priority of ‘the same invention’, referred to in Article 87(1) EPC, means that priority of a previous application in respect of a claim in a European patent application in accordance with Article 88 EPC is to be acknowledged only if the skilled person can derive the subject-matter of the claim directly and unambiguously, using common general knowledge, from the previous application as a whole.’
[153] The approach to be adopted was elaborated by this court in Unilin Beheer BV v Berry Floor NV [2004] EWCA (Civ) 1021, [2005] F.S.R. 6 at [48]:
“48 ... The approach is not formulaic: priority is a question about technical disclosure, explicit or implicit. Is there enough in the priority document to give the skilled man essentially the same information as forms the subject of the claim and enables him to work the invention in accordance with that claim.”
[154] In Abbott Laboratories Ltd v Evysio Medical Devices ULC [2008] EWHC 800 (Pat), [2008] R.P.C. 23, I added this:
“228. So the important thing is not the consistory clause or the claims of the priority document but whether the disclosure as a whole is enabling and effectively gives the skilled person what is in the claim whose priority is in question. I would add that it must “give” it directly and unambiguously. It is not sufficient that it may be an obvious development of what is disclosed.”
179 Having referred to those paragraphs of Kitchin LJ’s judgment in MedImmune, Floyd LJ (with whom Vos and Laws LJJ agreed) said in HTC Corp v Gemalto SA [2014] EWCA Civ 1335 at [65]:
[65] The skilled person must be able to derive the subject matter of the claim directly and unambiguously from the disclosure of the priority document. Mr Tappin stressed that the question was one of what was disclosed to the skilled person, not what was made obvious to him by the priority document, for example in the light of his common general knowledge. I agree that, as the above passage shows, that is the correct approach. That does not mean, however, that the priority document should be read in a vacuum. The question of what a document discloses to a skilled person takes account of the knowledge and background of that person. A document may mean one thing to an equity lawyer and another to a computer engineer, because each has a different background. The document still only has one meaning because it is only the relevant skilled person’s understanding which is relevant. What is not permissible is to go further than eliciting the explicit or implicit disclosure and take account of what a document might lead a skilled person to do or try, or what it might prompt him to think of.
180 Kitchin LJ (with whom Floyd and Longmore LJJ agreed) referred to that passage with approval in Icescape Ltd v Ice-World International BV [2019] FSR 5 at [42]. His Lordship then said at [43]:
[43] In my judgment the application of these principles provides a clear answer to the question before us. I accept that the key novel and inventive feature of the claimed invention is the use of joint elements (70) which enable the folding of the unit to take place. I am also prepared to accept that the skilled person would readily appreciate that one element of the kind depicted in figure 6 of the priority document could be joined to another to increase the width of the ice rink. But that does not alter the fact that there is no express or implicit disclosure in the priority document of two such elements joined together or of features A, D or E of claim 1 of the patent. Nor is it possible to derive these features directly and unambiguously, using common general knowledge, from the priority document as a whole. The skilled person seeking to implement the teaching of the priority document might join two elements together, but equally he might not, for he might have no need to do so. In these circumstances I have no doubt that Mr Alexander is inviting us to take a course which has been closed to us by the decision of the Enlarged Board in decision G2/98. It is not enough to say that features A, D and E do not relate to the function and effect, and hence to the character and nature, of the invention. The claim to priority depends upon the express or implicit disclosure of those features in the priority document and, since there is no such disclosure, the claim to priority must fail.
181 For the purposes of s 40(2)(a) it is necessary to identify the invention in the relevant claim of the complete specification and then ask whether there is a disclosure that is clear enough and complete enough for the invention to be performed by a person skilled in the art. Where the requirement of s 40(2)(a) is in issue, the claim and any related disclosure will be found within the one document (ie. the complete specification). However, where the requirement of s 43(2A)(b) is in issue, the sufficiency of the disclosure of the priority document will be assessed against the invention of the claim in the complete specification. Since that claim may not be reproduced in the priority document (which, as in the case of P1, might not include any claims at all) it is necessary to determine whether there is any disclosure of the invention in the claim in the priority document and, if so, whether it is clear enough and complete enough for it to be performed by the person skilled in the art.
182 ToolGen drew attention to the phrase “in respect of the same invention” in Art 87(1) of the EPC and the absence of those words in s 43(2A)(b) of the Act and submitted that s 43(2A)(b) imposed no requirement that the priority document and the patent application be in respect of the same invention.
183 The implication of ToolGen’s submission is that, under the Act, unlike the position under Art 87(1) of the EPC, a claim may be entitled to a priority date earlier in time than would otherwise be the case based on a prior application that is directed to an invention that is different from that in the claim. I do not think that is correct. In my opinion, the invention of the claim must be disclosed in the prior application and must be also disclosed in a manner that is clear enough and complete enough for the invention in the claim to be performed by the person skilled in the art.
184 ToolGen placed considerable reliance on the decision of Perram J in Encompass Corporation Pty Ltd v InfoTrack Pty Ltd (2018) 130 IPR 387 (“Encompass”). It submitted that Perram J’s decision was authority for the proposition that s 40(2)(a) does not require that the complete specification disclose the invention.
185 Encompass was concerned with s 40(2)(a) of the Act and the question whether the complete specification in suit (there was a number of them) complied with that provision. The question to which his Honour’s observations in that case were directed concerned a situation in which the description of the invention in a complete specification did not provide any explanation for the inclusion of a limitation in the relevant claim. The invention, as claimed, included a limitation which required a search “from at least one of a number of remote data sources”. As his Honour said at [152]:
[152] Section 40(2)(a) of the Act requires the specification to ‘disclose the invention in a manner which is clear enough and complete enough for the invention to be performed by a person skilled in the art’. The Respondent submitted that the defining characteristics of the invention as claimed was the requirement that the data sources to be accessed or queried should be ‘remote data sources’. Yet nothing in the specification of either Patent gave any hint as to why that limitation should exist. It was said that the invention variously described in the specifications was ‘database agnostic’.
His Honour continued at [155]:
[155] The specifications are, therefore, indifferent as to whether the data sources need to be remote or local. The invention described in them is unconcerned with issue of whether the data sources need to be remote. Yet the invention as claimed involves only a method which involves querying remote data sources. In that sense, the invention claimed is narrower than the invention disclosed. The narrowing is perhaps not very substantial given the very broad reading I would give the expression ‘remote data sources’. Nevertheless, I accept that what is claimed is not the same as what is described in the specification. As disclosed, the querying of ‘remote data sources’ is not an essential part of the invention yet the invention as claimed has this moderately limiting feature.
186 His Honour went on to note that, although the respondents agreed that the specifications were sufficient to enable a person skilled in the art to perform the invention, it submitted that s 40(2)(a) also required that the disclosure of the invention in the specification should be clear and that, in particular, s 40(2)(a) picked up the old “fair basis” requirement of a real and reasonably clear disclosure.
187 His Honour continued at [160]-[161]:
[160] The earlier form of s 40(2)(a) had, therefore, required that the invention be described fully. The concept of the ‘invention’ had been interpreted to mean ‘the embodiment which is described, and around which the claims are drawn’: Kimberly-Clark Australia Pty Ltd v Arico Trading International Pty Ltd (2001) 207 CLR 1; 177 ALR 460; 50 IPR 513; [2001] HCA 8 at [21]. The former requirement of s 40(2)(a) that the invention, in that sense, be fully described was interpreted as having two requirements:
(a) the specification had to make the nature of the invention plain to persons having reasonably competent knowledge of the subject; and
(b) it also had to make plain, to persons having reasonable skill, how to perform the invention.
(see Patent Gesellschaft AG v Saudi Livestock Transport and Trading Company (1997) 37 IPR 523 at 530 per Carr J, Jenkinson and Sackville JJ agreeing)
[161] The immediate question is whether that previous interpretation of the expression ‘describe the invention fully’ as including a requirement of making plain the nature of the invention to persons of reasonably competent knowledge of the subject has survived into the amended form of s 40(2)(a) which no longer includes that expression.
188 The question considered by his Honour was framed by him by reference to a judgment of the Full Court in Patent Gesellschaft AG v Saudi Livestock Transport and Trading Company (1997) 37 IPR 523 (“Patent Gesellschaft”) in which Carr J (with whom Jenkinson and Sackville JJ agreed) said when referring to s 40(2)(a) of the Act before amendment by the RTB Act at 530-531:
The specification contains a full description if it makes the nature of the invention plain to persons having reasonably competent knowledge of the subject and also makes it plain, to persons having reasonable skill, how to perform the invention: Edison & Swan United Electric Light Co v Holland (1889) 6 RPC 243 at 279; Samuel Taylor Pty Ltd v SA Brush Co Ltd (1950) 83 CLR 617 at 624-5. See also AMP Inc v Utilux Pty Ltd (1971) 45 ALJR 123 at 128 and Valensi v British Radio Corp Ltd [1973] RPC 337 at 377.
189 The respondents in Encompass contended that s 40(2)(a) of the Act required that the complete specification “make the nature of the invention plain” to persons skilled in the art. It is by no means clear what Carr J was referring to when using that phrase in Patent Gesellschaft. Certainly, s 40(2)(a) of the Act does not require that the specification identify the inventive step: see, for example, Winner v Ammar Holdings Pty Ltd (1993) 41 FCR 205 at 217 per Davis J (with whom Morling J agreed). I would also add that Patent Gesellschaft was decided before the High Court decided Lockwood Security Products Pty Ltd v Doric Products Pty Ltd (2004) 217 CLR 274 (“Lockwood”) and Kimberly-Clark Australia Pty Ltd v Arico Trading International Pty Ltd (2001) 207 CLR 1 (“Kimberly-Clark”). Lockwood does not make reference to the Full Court’s decision, and Kimberly-Clark only does so in support of the statement that s 40(2)(a) of the Act has never been construed to require that the body of the specification say what was essential to the invention. In my opinion, those decisions are inconsistent with there ever having been a requirement under s 40(2)(a) of the Act that the specification make “the nature of the invention plain” to persons skilled in the art.
190 In Encompass, Perram J rejected the argument that s 40(2)(a) of the Act as amended by the RTB Act required that the specification must make the nature of the invention plain. His Honour said at [163]:
… [T]he new form of s 40(2)(a) contains a requirement as to a clear disclosure but it is expressed to be ‘clear enough and complete enough’ for a particular purpose viz ‘for the invention to be performed by a person skilled in the relevant art’. I cannot see that any part of the wording of the new s 40(2)(a) which lends itself to being interpreted as containing a further requirement that the nature of the invention be made plain. Even if, which I do not accept, there was some peg in the language of s 40(2)(a) upon which such an interpretation might be hung, I do not see how it could be rescued from the purposive effect of the phrase ‘for the invention to be performed by a person skilled in the relevant art’. In short, s 40(2)(a) does not say as a matter of ordinary language that the specification must make the nature of the invention plain.
191 After referring to the Explanatory Memorandum to the RTB Act, Perram J observed at [165]-[167]:
[165] The Respondent submitted that removing the requirement of making the nature of the invention plain would hardly be raising the bar. There may be some rhetorical force in that flourish. Nevertheless, it seems to me that whilst the Explanatory Memorandum clearly demonstrates that its authors were aware of the former requirement of making the nature of the invention plain, it is silent on what they intended for the former requirement to continue. The entire discussion of the authors is given over to the second requirement of enablement. I think the better view is that the Exploratory Memorandum is silent on the topic of whether it was intended to remove the requirement to make the nature of the invention plain.
[166] … [T]here is accordingly no reason to depart from the language of s 40(2)(a) as it now stands.
[167] It follows that the only requirement now in s 40(2)(a) relates to enablement. Since the Respondent no longer advances a case that the specifications do not enable a person skilled in the relevant art to perform the invention, its case under s 40(2)(a) must fail.
192 His Honour’s decision is authority for the proposition that s 40(2)(a) does not require that the complete specification make the nature of the invention plain. In my opinion, his Honour is not to be taken as suggesting that there is no requirement under s 40(2)(a) that the invention be disclosed. Section 40(2)(a) expressly requires that the invention be disclosed in a manner which is clear enough and complete enough for the invention to be performed by the person skilled in the art.
193 Although neither s 43(2A) nor reg 3.13A(2) requires that the invention as claimed be “in respect of the same invention”, it does not follow that the priority document need not disclose the invention claimed. The invention claimed must be disclosed in the priority document if priority is to be obtained. It is not sufficient for the priority document to provide a starting point from which the person skilled in the art may transition from one invention to another by the use of the common general knowledge. The mere fact that it would be obvious to the person skilled in the art to use the disclosure in the priority document to produce what is claimed is not enough to obtain priority if, properly characterised, the priority document and the claim are for different inventions. In this respect, and notwithstanding differences between statutory language in the relevant UK and EPC provisions and s 40(2)(a) of the Act, I consider the position under Australian law is not materially different from the UK law as explained in the English authorities to which I have referred. The question is not what is made obvious to the person skilled in the art from the disclosure of the priority document but what it explicitly or implicitly discloses to that person. The distinction is important even though it is often not easily drawn.
P1 – DISCLOSURE AND ENABLEMENT
194 Section s 43(1) of the Act provides that each claim must have a priority date and different claims may have different priority dates. Section 43(3) of the Act provides that where a claim defines more than one invention, then, for the purpose of determining the priority date of the claim, it must be treated as if it were a separate claim for each form of the invention that is defined. Section 43(2A)(b) of the Act requires that there be a disclosure of “the invention in the claim” if a claim is to obtain priority in accordance with s 43(2A) of the Act.
195 For reasons previously stated, I do not accept ToolGen’s submission that s 43(2A)(b) does not require that there be a disclosure of the invention in the claim in the priority document. It follows from what I said that the invention claimed in claims 1 and 10 will only be entitled to priority if it is disclosed in P1. For this purpose, neither party drew a distinction between claim 1 and claim 10 and the submissions proceeded on the assumption that the priority date for claim 1 and claim 10 would be the same.
196 ToolGen submitted that the invention disclosed in P1 is an invention involving the use of:
(a) a Cas9 endonuclease (derived from a Type II CRISPR/Cas system) comprising a nuclear localisation sequence (NLS), which guides the Cas9 to the nucleus of a eukaryotic cell; and
(b) a single guide RNA comprising portions of CRISPR RNA (crRNA) and transactivating crRNA (tracrRNA) (also derived from the Type II CRISPR/Cas system),
to generate a site-specific double stranded break in the DNA of a eukaryotic cell at a target sequence adjacent to a protospacer adjacent motif (PAM).
197 I should say at once that there is a difficulty with ToolGen’s formulation of the invention in these terms. While it may be accepted that the invention disclosed in P1 involves the use of a Cas9 endonuclease and a single guide RNA of the kind described by ToolGen, it does not follow that it is the invention disclosed.
198 The same comment may be made in relation to the Abstract in P1 which refers to “a novel genome editing technology based on RNA-guided Cas9 endonucleases” which language is repeated in the first paragraph of the section of P1 headed Main Text. That does not constitute a disclosure of any genome editing technology based on RNA-guided Cas9 endonucleases. The Abstract is no more than a general introduction to what is disclosed in P1 and does not disclose the invention of the claims. It is an introductory statement that provides context for what follows.
A nucleic acid encoding a guide RNA
199 The respondents’ case on this issue is focused on the method of introducing the nucleic acid encoding the guide RNA into the eukaryotic cell as required in claim 10, given that there is no express requirement in claim 1. On the use of plasmid DNA encoding the guide RNA as a means of introducing the guide RNA into the cell, Associate Professor Firestein accepted that P1 does not disclose the use of plasmid DNA encoding either of the single guide RNAs that it discloses. Professor Thomas and Associate Professor Herold agreed. Associate Professor Firestein also accepted that the particular CRISPR/Cas9 system disclosed in P1 involves the use of in vitro transcribed guide RNA and does not disclose the use of plasmid DNA encoding the guide RNA as a means of introducing into the cell the guide RNA. Associate Professor Firestein said:
It does not disclose that, but to me this would be a obvious thing that could be done based on my experience with expressing similarly sized RNAs via plasmid encoded DNA.
200 Under the heading the “Summary of the Invention” appears a reference to “nucleic acid encoding RNA-guided Cas9 endonucleases”. Associate Professor Firestein considered that the phrase “RNA-guided Cas9 endonucleases” referred to both the guide RNA and the endonuclease or, in other words, the complex which is formed in the cell. ToolGen contended that the reference to RNA-guided Cas9 endonucleases is a broad disclosure that is ambiguously agnostic as to how the guide RNA is made and therefore extends to plasmid DNA that is transcribed into guide RNA in the eukaryotic cell. ToolGen submitted, in effect, that the language used in the Summary of Invention was, deliberately broad, and should not be read down by reference to what is more specifically described elsewhere in P1, including by reference to the experiments conducted which did not use plasmid encoded guide RNA.
201 Associate Professor Herold considered that the reference in the Summary of the Invention to “nucleic acids encoding RNA-guided Cas9 endonucleases” is referring only to the Cas9 endonuclease which is encoded by plasmid DNA introduced into the cell. On this interpretation of the relevant phrase, the word “RNA-guided” is an adjective that describes the Cas9 endonuclease that is encoded rather than the guide RNA and Cas9 endonuclease individually. This is not the interpretation that was adopted by Professor Thomas, but it is fair to say that he agreed that Associate Professor Herold’s view was a reasonable one. Professor Thomas gave the following evidence:
MR CORDINER: Yes. Now, I put it to you, if the word or term “RNA guided Cas9 nucleases” is being used as it’s used almost – well, used everywhere else in P1, it means the guide RNA and the endonuclease; do you agree with that?
PROF THOMAS: As I said before, I have interpreted it in that fashion, in terms of it being the complex, that’s true, but you could also construe it as being an RNA guided endonuclease, in other words, referring to Cas9.
MR CORDINER: And if we’re talking about the complex, if you’re describing, here, an embodiment of the invention – sorry, some embodiment of the present invention to providing nucleic acids encoding the complex, if I put it that way, a reasonable position to take would be it’s describing, there, using nucleic acids to encode the guide RNA and to encode the Cas9; could you agree with that?
PROF THOMAS: Well, that’s a difficult one for me to answer in the sense that I was asked to interpret P1 in the context of the entire document.
MR CORDINER: Yes.
PROF THOMAS: And I cannot see any evidence in this document for, for example, a DNA encoding an – the RNA component of the complex. So to me, the most correct interpretation of this – this phrase, within the summary of the Summary of the Invention, is that it refers to a nucleic acid, let’s say DNA, encoding the RNA guided Cas9 endonuclease. To me, that’s the most straightforward interpretation when I look at the document as a whole.
MR CORDINER: And when you’re looking at the document as a whole, the document itself doesn’t, at any point, say you can’t use plasmid encoded guide RNA, does it.
PROF THOMAS: It makes no reference to plasmid encoded guide RNA.
MR CORDINER: So it doesn’t say you can’t use plasmid encoded guide RNA, does it.
PROF THOMAS: It doesn’t say that it can’t be used.
202 Associate Professor Herold’s interpretation is consistent with what is disclosed in P1. The Cas9 protein is produced using “Cas9-encoding plasmids” whereby DNA encoding the relevant protein is introduced into the cell using a plasmid. However, in each of the two working examples described in P1, the guide RNA is in vitro transcribed RNA and is introduced into the cell in naked or isolated form (and without the use of a plasmid). There are various other statements in P1 which tend to support Associate Professor Herold’s interpretation of the Summary of the Invention.
203 First, the Abstract makes reference to “synthetic guide RNAs”. As Professor Thomas explained, this is a reference to the preparation of the guide RNA by a synthetic means using ribonucleotides (a nucleotide containing ribose) or in vitro transcription in which the RNA polymerase enzyme opens a double-stranded DNA and one strand of the exposed nucleotides is used as a template for the synthesis of RNA. The phrase “synthetic guide RNA” does not describe RNA transcribed from DNA in vivo. Associate Professor Firestein agreed that a guide RNA which is produced in a cell by transcription using plasmid DNA which encodes the guide RNA it is not accurately described as a synthetic guide RNA.
204 Secondly, the Abstract makes clear that RGENS are customised without any cloning step (ie. without inserting a DNA fragment into a vector). In Thomas 1, Professor Thomas referred to some of the difficulties associated with the use of ZFNs and TALENs in genome editing which, because they are proteins, are difficult to introduce into the cell. To overcome this difficulty, molecular cloning techniques were developed to make circular DNA constructs that encode ZFNs and TALENs that were then introduced into the cell. In relation to the guide RNA, Professor Thomas explained that the advantage of having a system that could be customised without any cloning step means that the target location for DNA cleavage provided by the guide RNA in the Cas9 system can be changed by synthesising a new guide RNA without the need to clone nucleotide sequences into vectors such as plasmids. This means that different DNA sequences can be targeted by changing the guide RNA. From Professor Thomas’ evidence on this topic, which I accept, it is apparent that, at least as presented in the Abstract, a significant advantage of the genome editing technology described in P1 is that it uses synthetic guide RNAs that may be customised without any cloning step and, in the words of the authors, providing a “… broadly useful, scalable and expeditious platform for genome engineering in cells and organisms”.
205 It was suggested by Associate Professor Firestein in his evidence that the in vitro transcription templates appearing at page 11 of P1 were templates from which DNA encoding the guide RNA could be constructed for in vivo use. He gave evidence that the DNA templates could be cloned into a plasmid in vitro and then introduced into the cell to transcribe RNA in vivo. He estimated that this would take approximately one week to do as his laboratory would have had the plasmids available. He also gave evidence of an alternative method whereby the DNA templates could be introduced into the cell to transcribe RNA in vivo. He did not give evidence of how long this would take. He acknowledged that both approaches would only work if the promoters were changed from a T7 promoter to a U6 or H1 promoter (which are eukaryotic promoters).
206 Associate Professor Herold gave evidence that cloning the DNA template into a plasmid would definitely take longer than a week but could be done. Professor Thomas gave evidence that using a linear DNA strand in vivo was, in his opinion, not an approach that anyone would routinely use because of concerns that it might degrade in the cell and that it is much easier to generate a circular plasmid which is more stable in the cell. He also agreed that the promoters would need to be changed for both approaches and said that this would take several weeks to validate. I think the significance of this evidence is that P1 does not disclose a DNA template for a guide RNA suitable for in vivo use. Rather, what is disclosed are two DNA templates suitable for in vitro preparation of a guide RNA.
207 I accept that it would not be a difficult exercise for a molecular biologist in possession of the information in P1 coupled with the common general knowledge to use a plasmid DNA encoding the guide RNA to produce the guide RNA in vivo using standard techniques that were well known at the priority date. I also accept that it would be obvious to the skilled addressee that he or she could use plasmid DNA encoding a guide RNA as a means of generating the guide RNA in the cell. On this particular point, I accept Associate Professor Firestein’s evidence.
208 I also accept, as did all three expert witnesses who gave evidence on this topic, that P1 does not exclude the use of plasmid DNA encoding the guide RNA in place of naked RNA created in vitro. However, to say that this is not excluded does not mean that it is disclosed. P1 makes no reference to plasmid DNA encoding a guide RNA.
209 Further, P1 does not disclose the use of plasmid DNA encoding a guide RNA as a means of introducing a guide RNA into the cell. P1 is directed to the use of a guide RNA produced in vitro (ie. naked or isolated RNA) which is then introduced into the cell. There is no disclosure of any system in which DNA (or viral RNA) is introduced into the cell in order to transcribe the guide RNA in vivo.
210 With regard to the term RNA-guided endonucleases in the Summary of the Invention, the language used is in my opinion ambiguous. But if the Summary of the Invention is read in the context of P1 as a whole, I think the notional skilled addressee would, like Associate Professor Herold, understand the term as referring to the Cas9 endonucleases which are elsewhere described as having been introduced into the cell by means of a Cas9 encoding plasmid. In this regard, I prefer the evidence of Associate Professor Herold to that of Associate Professor Firestein and, to the extent that it was inconsistent with Associate Professor Herold’s evidence on this point, the evidence of Professor Thomas.
211 Even if I am wrong about this, there is another way of understanding the Summary of the Invention that avoids any redundancy in the use of the words “vectors comprising Cas9 endonucleases” which immediately follow the words “RNA-guided Cas9 endonucleases” and which is consistent with what is more specifically described in P1. As I have explained, P1 does describe nucleic acid encoding guide RNA in the transcription templates. However, these are DNA templates for encoding guide RNAs in vitro. In those circumstances, it makes sense to interpret the phrase “nucleic acids encoding RNA-guided Cas9 endonucleases” as referring to nucleic acids encoding the guide RNAs in vitro (as shown in P1) and nucleic acids encoding the Cas9 endonuclease in vivo using a vector (as shown in P1). This construction of the Summary of the Invention avoids any redundancy arising from the presence of the words “vectors comprising Cas9 endonucleases” which, as ToolGen submitted, arises on Associate Professor Herold’s interpretation. In any event, whether or not the relevant phrase is construed as referring to nucleic acids encoding an endonuclease or nucleic acids encoding both the guide RNA and the endonuclease, the Summary of the Invention does not encompass the use of nucleic acids encoding a guide RNA in the cell, such as through the use of plasmid DNA introduced into the cell.
212 Claims 1 and 10 (and, with the exception of claim 19, the dependent claims) are directed to an invention in which the guide RNA of the claims is introduced into the cell in the form of nucleic acid (DNA or viral RNA) which then encodes the guide RNA in the eukaryotic cell. P1 does not disclose any such system either explicitly or implicitly. It follows that those claims are not entitled to priority based on P1.
A Type II CRSIPR/Cas system from a bacterial species other than S. pyogenes
213 The next question is whether P1 discloses a system for cleaving DNA using a Cas9 polypeptide derived from a bacterial species other than S. pyogenes in a manner which is clear enough and complete enough for the invention of the claims to be performed by a person skilled in the art. Although some of the dependent claims are limited to a Cas9 polypeptide derived from S. pyogenes, none of claims 1, 2, 10 or 11 are so limited.
214 It is common ground that P1 discloses a CRISPR/Cas9 system derived from S. pyogenes. However, ToolGen submitted that the disclosure of the bacterial species from which the Cas9 endonuclease is derived is not limited to S. pyogenes, and that the only relevant limitation is that the Cas9 comes from a Type II CRISPR system capable of forming an active endonuclease when complexed with a guide RNA. In this regard, ToolGen placed emphasis upon the following express disclosure in P1:
“Cas9, an essential protein component in the Type II CRISPR/Cas system, forms an active endonuclease when complexed with two RNAs termed CRISPR RNA (crRNA) and transactivating crRNA (tracrRNA), thereby slicing foreign genetic elements in invading phages or plasmids to protect the host cells.”
However, that passage is describing Cas9 in the Type II CRISPR/Cas system in bacteria where it provides immunity against invading phages and plasmids. It is background information which assists the reader’s understanding of the mechanism of action of the invention disclosed in P1. It is not a disclosure of the invention.
215 ToolGen also drew attention to the use of the word “systems” as appearing in the Abstract. It submitted that word indicates that the authors are not merely referring to the S. pyogenes system, but to all Type II CRISPR/Cas systems. Reliance also was placed on the statements in P1 (at page 5) concerning the limitations of the requirement for a 5’-GG-3’ dinucleotide in the PAM sequence and the suggestion that “[t]his limitation might be relieved by engineering Cas9 or employing Cas9 derived from another species”. Here again, the use of the word “systems” occurs in the context of a description of the background to the technology including the role such systems play in protecting prokaryotes against invading phages and plasmids.
216 ToolGen also submitted that the disclosure in P1 of PAMs which Type II Cas9 endonucleases recognise is not limited to those derived from S. pyogenes, but also encompass any PAM that is recognised by a Type II Cas9 protein, and that this set of PAMs, as it described them, is implicitly disclosed by P1 because it discloses that “Cas9 is a sequence-specific endonuclease in Type II CRISPR systems”. ToolGen submitted that the disclosure of P1 extends to Cas9 from all bacterial species which have a Type II CRISPR system, provided that the Cas9 is used in combination with a PAM that it recognises. It further submitted that there is no requirement for P1 to disclose the nucleotide sequences for all such PAMs or all such Cas9s polypeptides.
217 In essence, ToolGen’s argument is that P1 discloses a system in which any Type II Cas9 polypeptide that recognises any known PAM (including but not limited to 5’-NGG-3’) may be used. I do not accept that any of the claims of the patent application are limited in their scope to the use of a Type II Cas9 polypeptide that recognises any known PAM. Even if they were to be construed as limited to the use of a Type II Cas9 polypeptide that recognises a known PAM sequence, that would not mean that the use of any Type II Cas9 was sufficiently enabled. This is because P1 does not identify any principle of general application which would permit the skilled addressee to determine whether any particular Type II Cas9 might reasonably be expected to work.
218 A similar argument to that relied on by ToolGen was considered by Kitchin J (as his Lordship then was) in Novartis AG v Johnson & Johnson Medical Ltd [2009] EWHC 1671. His Lordship said at [244]:
It follows, in my judgment, that a claim to a class of products said to possess a useful activity must be based upon the identification of a common principle which permits a reasonable prediction to be made that substantially all the claimed products do indeed share that activity. Further, it is not permissible to by-pass that requirement simply by adding a functional limitation which restricts the scope of the claim to all the products which do have the relevant activity, that is to say all those which “work”. In the case of a claim limited by function, it must still be possible to perform the invention across the scope of the scope of the claim without undue effort. That will involve a question of degree and depend upon all the circumstances including the nature of the invention and the art in which it is made. Such circumstances may include a consideration of whether the claims embrace products other than those specifically described for achieving the claimed purpose and, if they do, what those other products may be and how easily they may be found or made; whether it is possible to make a reasonable prediction as to whether any particular product satisfies the requirements of the claims; and the nature and extent of any testing which must be carried out to confirm any such prediction.
219 I respectfully agree with his Lordship that if a claim is limited by a functional requirement it may still be necessary to consider whether there is an undue burden involved in identifying whether embodiments not specifically described in the specification or, in this case, the priority document, meet the functional requirement. It is not necessary to consider the matter further because, in my view, none of the claims include any such functional requirement.
220 Senior Counsel for the respondents put to Associate Professor Firestein by that P1 does not disclose a system from any other bacterial species (apart from S. pyogenes) that can mediate DNA cleavage in the eukaryotic cell. He said that it did not show that directly, but that it does not exclude the possibility of that occurring. When pressed on this topic, he said:
I will just be very exact and say that P1 does not show any experiments or provides [sic] any direct information on another CRISPR-Cas9 system, but it does disclose the existence of other CRISPR-Cas9 systems, and raises the potential for employing these other Cas9 systems for similar purposes.
In Firestein 1 (which he cross-referenced in JER 1) Associate Professor Firestein referred to the statement in P1 (at page 5) that engineering Cas9 or employing Cas9 derived from other species might relieve the limitation arising out of the requirement for a 5’-GG-3’ dinucleotide when using a Cas9 protein derived from S. pyogenes. He said that this contemplates the use of other Cas9 proteins.
221 I accept that P1 discloses, in a general sense, the existence of Cas9 proteins derived from other bacterial species and the possibility that they may be used to mediate DNA cleavage in eukaryotic cells. But what is clear from P1 is that this is raised by the authors as a mere possibility. P1 does not include any further discussion of this possibility nor does it present any evidence or commentary from which it may be inferred that all, or even some, Type II Cas9 proteins derived from other bacterial species could reasonably be expected to work with either the 5’-GG-3’ PAM or other PAMs to mediate DNA cleavage in eukaryotic cells.
222 Associate Professor Firestein went on to say that P1 was a “starting point” or a “springboard for potentially exploring their use in a similar assay”. He agreed that P1 does not disclose which species of bacteria (apart from S. pyogenes) have a Type II system or which of the different Type II systems may work in eukaryotes. He also agreed that it does not disclose what Type II systems may recognise a non-NGG PAM. Professor Thomas and Associate Professor Herold agreed that P1 does not show CRISPR/Cas9 components from any bacterial species other than S. pyogenes. This includes not only the polypeptide, but also the guide RNA and its components. They also agreed that P1 does not disclose which other species of bacteria or archaea have a Type II system or whether any would work in eukaryotes.
223 In Firestein 1, Associate Professor Firestein said that, in his opinion, P1 does not exclude the possibility that S.pyogenes CRISPR/Cas9 system may utilise non-NGG PAMs. Although Associate Professor Firestein suggested in his oral evidence that Cas9 derived from S. pyogenes might recognise other PAM sequences apart from 5’-NGG-3’ and that this is something he would investigate, he did not suggest this was disclosed in P1. Professor Thomas and Associate Professor Herold were clear that P1 does not disclose the use of a Cas9 polypeptide which recognises PAM sequences other than those recognised by S. pyogenes derived Cas9.
224 I do not accept ToolGen’s argument that P1 provides a broad disclosure of any Type II Cas9 polypeptide that recognises any PAM sequence or any known PAM sequence. Rather, P1 suggests that the use of Cas9 derived from other species might enable other PAM sequences to be used. This accords with Associate Professor Firestein’s evidence that P1 provides a starting point from which to explore such possibilities.
225 I previously found that P1 does not incorporate Jinek by reference. However, even if P1 is considered together with Jinek there is still no disclosure of a system using a Cas9 polypeptide derived from a bacterial species other than S. pyogenes. Although Jinek provides sequences for other Cas9 orthologs (sometimes spelt orthologues) (including S. thermophilus and L. innocua), they are only shown to work with a 5’-NGG-3’ PAM in vitro in prokaryotes. Jinek did not test their ability to recognise a non-NGG PAM and did not identify a non-NGG PAM recognised by Cas9 from any other bacterial species.
226 Moreover, the chimeric sgRNA disclosed in Jinek is a S. pyogenes derived chimeric sgRNA (ie. the same as disclosed in P1). Associate Professor Firestein gave evidence that he would use the information in Jinek to test the Cas9 orthologs identified in the paper (L. innocua and S. thermophilus Cas9) in combination with the chimeric guide RNA from S. pyogenes for their ability to mediate CRISPR/Cas9 DNA at non-NGG PAM sites. Even if that were considered an obvious thing to do, that does not broaden the disclosure of P1 to encompass a system for cleaving DNA using a Cas9 polypeptide derived from other bacterial species.
227 I find that P1 does not disclose an invention in which the Cas9 protein is derived from any species of bacteria other than S. pyogenes. Nor does it disclose an invention which encompasses the use of any Cas9 which is shown to recognise a non 5’-NGG-3’ PAM. However, if I am wrong about that and it does make such a disclosure, the next question is whether that disclosure is clear enough, and complete enough, for the invention to be performed by the person skilled in the relevant art.
228 The respondents submitted that even if P1 does disclose an invention the components of which are derived from bacterial species that are not limited to S. pyogenes, the skilled addressee is not enabled by P1 to work the invention across the breadth of the claims. They submitted that P1 does not provide any meaningful guidance as to how the work required to perform the invention across the breadth of the claims could be done. They further submitted that any such work would not be routine, and would impose an undue burden on the person skilled in the art by requiring them to undertake a considerable amount of work in circumstances where it would not have been clear that such a research project would ultimately succeed.
229 The respondents submitted that the work involved in developing another Type II CRISPR/Cas system would include at least:
(a) identifying a Cas9 nuclease from another bacterial species and how to localise it to the nucleus of a eukaryotic cell;
(b) identifying the endogenous crRNA direct repeat and spacer sequences associated with it;
(c) identifying what portion of a direct repeat sequence is required for a mature crRNA molecule;
(d) identifying the endogenous tracrRNA sequence associated with the Cas9 nuclease;
(e) identifying and validating the PAM site recognised by the Cas9 nuclease in eukaryotic cells;
(f) identifying and validating the length of the target nucleic acid sequence that can be cleaved by the Cas9 nuclease;
(g) identifying which portions of the mature crRNA molecule and tracrRNA sequence are required for the crRNA and tracrRNA portions of a sgRNA;
(h) designing a sgRNA that successfully guides the Cas9 to effect DNA cleavage in eukaryotic cells; and
(i) investigating whether system can effect cleavage in a eukaryotic cell.
230 ToolGen contended that the respondents’ summary of the tasks that would need to be performed by the skilled addressed involved “… atomising the task which must be performed to as final level of granularity as possible, so as to multiple them …”. However, ToolGen acknowledged that, in substance, the main tasks involved in performing the invention of the claims using Cas9 derived from species other than S. pyogenes involved the following main tasks:
(a) identifying another bacterial species with a Type II CRISPR system;
(b) determining the endogenous crRNA and tracrRNA sequences;
(c) characterising the mature crRNA and tracrRNA molecules endpoints and lengths;
(d) ascertaining the PAM sequence which the Cas9 of that bacterial species recognises; and
(e) combining the crRNA and a portion of the tracrRNA into a single chimeric guide RNA using the methods disclosed in Jinek.
231 ToolGen also submitted that it is not necessary for P1 to disclose all of the bacterial species that could be used to perform the invention. It submitted that the sequences of the Cas9 polypeptide, mature crRNA and mature tracrRNA (including knowledge as to the PAM recognised by the Cas9) are all “input elements” which are not required to be available at the priority date for enablement across the whole scope of the claims. It further submitted that all that P1 and the patent application must enable is how those input elements are combined and that such combinations become available to be used in the claims as and when they become known. ToolGen relied on a decision of the Technical Board of Appeal in, Genentech I/Polypeptide expression (T292/85) 27 January 1988 (“Polypeptide”) where the Board stated at [3.1.3]:
What is also important in the present case is the irrelevancy of the particular choice of a variant within the functional terms ‘bacteria’, ‘regulon’ or ‘plasmid’. It is not just that some result within the range of polypeptides is obtained in each case but it is the same polypeptide which is expressed, independent of the choice of these means. A term of this kind must, of course, be clear and enable the skilled person to find suitable specimens without undue difficulty. In the present application enough choice is available, although some vehicles and hosts are preferred for practical reasons.
232 The reasoning in the Polypeptide case does not apply here. There is no evidence presented in P1 to suggest that all Type II CRISPR/Cas9 systems can cleave DNA in eukaryotic cells. Nor is there evidence presented in P1 to suggest that the Cas9 polypeptide in S. pyogenes is the same as that found in other bacteria with a Type II CRISPR/Cas system. That the Cas9 polypeptides will vary between species is apparent from P1 itself which recognises that different species may recognise different PAMs. P1 does not disclose any principle of general application from which it would appear reasonably likely that use of all species within the scope of the claims (whether known or yet to be discovered) are enabled: Regeneron at [56] (vi)-(viii).
233 The question whether P1 enables the invention of claim 1 and 10 to be performed across their breadth depends at least to some extent, on whether, as I have found, the skilled addressee is a molecular biologist or, as ToolGen submitted, a skilled team comprising a molecular biologist and a microbiologist. In ToolGen’s submissions Professor Giffard was identified as a microbiologist with expertise in bacterial CRISPR/Cas systems and that a person such as him would form part of the skilled team. For reasons previously explained, I do not accept that submission.
234 ToolGen’s analysis of how the skilled team would proceed to develop a system using another bacterial species drew heavily on the evidence of Professor Giffard and certain papers which he identified (Bhaya (2011), Makarova (2011) and Deltcheva (2011)) which ToolGen submitted were common general knowledge of microbiologists as at the priority date. It also made submissions which assumed that one or more of those papers would have been ascertained by a molecular biologist working in the field of gene editing in eukaryotic cells by means of a literature search. In this way the papers identified by Professor Giffard were said by ToolGen to be relevant to sufficiency even if the microbiologist was not a member of the skilled team.
235 The evidence of the molecular biologists, in particular, Associate Professor Herold, establishes that Bhaya (2011), Makarova (2011) and Deltcheva (2011) were not common general knowledge of molecular biologists working in the field of gene editing in eukaryotic cells at the priority date. I make the same finding in relation to the CRISPRFinder tool referred to in Professor Giffard’s evidence based on Associate Professor Herold’s evidence that this was not something he was familiar with in October 2012.
236 As to Professor Giffard, he was cross-examined extensively on his knowledge of Makarova (2011) and Deltcheva (2011) as at the priority date. Although he was unable to be certain whether or not he had read Makarova (2011) and Deltcheva (2011) prior to October 2012, I accept his evidence that it is likely that he did so. I also accept his evidence that, as at March 2012 when he referred to Bhaya (2011) in a document he prepared in connection with a funding submission, it was a key publication in the field. Bhaya (2011) was a review article which discussed the question of the proper classification of CRISPR systems and, in this context, referred to the paper by Makarova et al published in Nature Reviews Microbiology in June 2011 and the paper by Deltcheva et al published in Nature in March 2011. In my view, it is likely these were significant publications in the field as at October 2012. Professor Giffard considered the journals in which they were published to be prominent and prestigious and he regarded each of the three papers as important in either summarising or advancing research into CRISPR/Cas systems in bacteria and archaea at the time of publication. He considered them essential reading for a microbiologist wishing to have an understanding of such systems. I accept that evidence. I note that both Makarova (2011) and Deltcheva (2011) were cited in Bhaya (2011) and that both Bhaya (2011) and Deltcheva (2011) are cited in Jinek. I accept that Bhaya (2011), Makarova (2011) and Deltcheva (2011) were common general knowledge of a microbiologist working in the CRISPR/Cas field in October 2012. I should note that Professor Giffard did not give any evidence to suggest that he was familiar with Jinek as at the priority date.
237 ToolGen submitted that Makarova (2011) and Deltcheva (2011) were relevant not only as common general knowledge of a microbiologist, but also as information likely to have been ascertained by a molecular biologist working in the field of gene editing in eukaryotic cells who was seeking to carry out what ToolGen says is the invention disclosed by P1 using Cas9 derived from bacterial species other than S. pyogenes. ToolGen submitted that there was nothing wrong in principle in having regard to information obtained by means of a literature search provided that it could not be said that the exercise imposed an undue burden.
238 The authorities are clear that the question of sufficiency is to be ascertained by reference to the scope of the relevant disclosure to a person skilled in the art armed with the relevant common general knowledge. Unless the skilled addressee is directed to another document that discloses additional information or which forms part of the skilled addressee’s common general knowledge, it is outside the scope of information that may be taken into account when considering whether or not there is an enabling disclosure. The test is whether the invention can be performed using information disclosed by the relevant publication coupled with the common general knowledge without imposing an undue burden. Moreover, the fact that it is necessary for a worker in the field to utilise scientific literature that is not common general knowledge (especially if it is from a different field) in order to perform the invention would suggest that there is no enabling disclosure: Gilead at [221].
Identifying another bacterial species with a Type II CRISPR/Cas System
239 There are a number of approaches which ToolGen submitted the skilled addressee could have taken, as at the priority date, in order to identify other bacterial species with endogenous Type II CRISPR/Cas9 systems that could be used to perform the invention of the claims without undue burden. One such approach was laid out in the evidence of Professor Giffard, assuming he is a reasonable proxy for a microbiologist with expertise in the field of bacterial CRISPR/Cas9 systems in October 2012 and who would form part of a skilled team. It involved using Makarova (2011) (and particularly the information in the supplementary table S1) to identify a bacterial species with a Type II CRISPR system. This would have provided the microbiologist with a non-exhaustive list of approximately 120 bacterial species (including S. pyogenes and S. thermophilus) with Type II CRISPR systems.
240 Professor Giffard also adopted an alternative approach to identifying other bacterial species with an endogenous Type II CRISPR/Cas system. It involved conducting a search of the NCBI genome sequence database known as Genebank maintained by the National Institutes of Health. This database was a standard research tool used by scientific researchers in October 2012. Associate Professor Herold agreed that different specialities within the molecular biology field used different tools and that NCBI was a standard tool used by all molecular biologists. Professor Giffard used it at the time of preparing his evidence to search for nucleotide sequences that were annotated as coding for the Cas9 or Csn1 protein (the latter being an earlier name used to describe Cas9). He refined his search to results limited to bacteria and those that were released between 1 January 1900 and 30 September 2012. According to his evidence, 1,513 results were generated, the second of which was identified as “streptococcus thermophilus MS-ZLW-002”.
241 Upon inspecting the NCBI search result, Professor Giffard observed that it included an annotation indicating that the sequence included a gene encoding for Csn1/Cas 9 and he was able to locate the position of this gene. He was also able to identify the genes encoding for Cas 1 and Cas 2 through annotations but he was not able to identify the CRISPR array as there was no annotation identifying it.
242 Using the genomic sequence obtained for S. thermophilus MS-ZLW-002, Professor Giffard then used the CRISPRFinder tool to identify the DNA sequences for the CRISPR array and its location in the bacterial genome. He conducted his search by copying and pasting the genome sequence of S. thermophilus MS-ZLW-002 from the NCBI database into CRISPRFinder and searching for a CRISPR array within the sequence. The results of the search showed two CRISPR arrays in different positions. The second CRISPR array was 2000 base pairs from the gene for Csn1/Cas9 which he considered to be “extremely close”. He therefore concluded that this was a component of the same Type II CRISPR/Cas locus as the genes for Csn1/Cas9, Cas1 and Cas2. By this means he was able to identify the CRISPR array comprising the repeat sequences (shown in yellow to the left in the figure below) and the spacer sequences (shown in various colours to the right in the figure below).
243 According to Professor Giffard, if he was able to identify Cas1, Cas2, Cas9/Csn1 and the CRISPR array within reasonably close proximity of each other within the bacterial genome, he would have a high degree of confidence that he had identified the Type II CRISPR/Cas system of the bacterial genome in question. He considered it would not be necessary to also identify the other Cas associated proteins, Cas4 or Csn2, since, in his experience, the identification of the other components provides an extremely reliable identification of a functional CRISPR system. I accept that evidence which in my opinion is likely to reflect the use by Professor Giffard of the common general knowledge and the ordinary skill of a microbiologist working in the field of CRISP/Cas systems at the priority date.
244 It may be observed that Professor Giffard’s exercise identified one bacterial species with a Type II CRISPR system, ie. S. thermophilus. The evidence shows that the Type II CRISPR system of S. thermophilus recognises a 5’-NGG-3’ PAM when used in vitro. A post priority date paper by Fonfara et al (“Fonfara (2013)”) published in late 2013 indicates that S. thermophilus recognises an NGGNG PAM sequence (which encompasses NGG) in vivo. Accordingly, S. thermophilus does not overcome the limitation of S. pyogenes referred to at page 5 of P1.
245 Associate Professor Firestein also proposed an alternative approach to identify and investigate Cas9 orthologs which involved conducting a homology search using the S. pyogenes Cas9 sequence as the reference sequence to identify genes in bacteria that show a high degree of homology. He said that he would undertake a homology analysis because prior to October 2012 he understood that enzymes that are highly homologous are likely to work by the same or a similar mechanism of action. Given this, he reasoned that it was logical that two Cas9 orthologs, when highly homologous to each other, may exhibit similar activity.
246 Associate Professor Firestein gave evidence that he would undertake the homology search either at the level of DNA using NCBI BLAST or at the level of amino acids using UniProt, which were both databases routinely used by him before October 2012. He said he would use the search results to assess the degree of sequence homology across the Cas9 gene and would select Cas9 orthologs that have conservation in specific effector domains (eg. endonuclease domains) and homology (of at least 50-60%) across the entire sequence.
Determining the endogenous crRNA and tracrRNA sequences
247 The next step identified by ToolGen that the skilled addressee would need to take to perform the invention of the claims in other Type II CRISPR-Cas bacterial species was to identify the endogenous crRNA and tracrRNA sequences.
248 In Giffard 1, Professor Giffard stated that he understood from his own work prior to October 2012 that crRNA arises from the transcription of the CRISPR array into a single pre-crRNA molecule, which is then processed into individual crRNA molecules by RNA cleavage events. He also understood that each crRNA molecule contained a transcribed spacer sequence. He therefore understood that the nucleotide sequence comprising the CRISPR array is the DNA coding for crRNA. Given that he had already identified the nucleotide sequence and location of the CRISPR array through his search on the CRISPRFinder, he was able to determine the DNA sequence coding for crRNA in S. thermophilus MS-ZLW-002.
249 When it came to determining the DNA sequence coding for tracrRNA, Professor Giffard relied on the Bhaya (2011) which stated that:
It was also recently established that a trans-encoded small CRISPR RNA (tracrRNA) is involved in the processing of pre-crRNA into crRNA in Type II systems through the formation of a duplex with the CRISPR repeat sequence.
He stated that he understood the expression “formation of a duplex” to mean that tracrRNA can associate with the repeat-derived RNA by base pairing. This means that the tracrRNA can bind with the parts of the single stranded pre-crRNA that are transcribed from the repeat sequences of the CRISPR array (recalling that pre-crRNA is comprised of both transcribed repeat and spacer sequences). This understanding is confirmed by another passage in Jinek which ToolGen relied on and which states:
[T]ype II systems process pre-crRNAs by different mechanisms in which a trans-activating crRNA (tracrRNA) complementary to the repeat sequences in the pre-crRNA triggers processing.
250 ToolGen submitted that the implication of this statement is that the DNA sequence comprising the repeat sequence in the CRISPR array is complementary to the nucleotides comprising tracrRNA and must therefore be identical to the DNA sequence that codes for tracrRNA. Therefore, using the repeat sequence obtained from the CRISPRFinder search, Professor Giffard could identify the DNA sequence that would be the same nucleotide sequence as the DNA sequence coding for tracrRNA.
251 In oral evidence, Professor Thomas gave evidence that appeared to confirm Professor Giffard’s understanding:
MR FLYNN: Yes. Now, if you take that as your identification of the DNA sequence that corresponds to the type 2 Crispr/Cas system, could you explain your comment in the joint report that the DNA sequence encompassing the sequence specifies Crispr RNA and tracrRNA would be available?
PROF THOMAS: Yes. Although I wasn’t referring specifically to that sequence. You would understand of course. But as a general point, what I was trying to communicate there was that the equivalent system for the hypothetical type 2 Crispr/Cas system, for that particular bacteria, would have the arrangement of spacers and then repeat sequences in a similar manner to what’s shown in that figure [referring to the CRISPRFinder figure].
MR FLYNN: Yes. And then, if you had the spacer and the repeat sequences [from the CRISPRFinder figure], then how do you get from that to the DNA sequence encompassing the sequence specifying firstly, the Crispr RNA?
PROF THOMAS: Well, I know from my knowledge of the system that there are two parts to the Crispr RNA. There’s the part that binds to the pathogen and there’s a repeat spacer sequence as well, which binds to the tracrRNA. So, the unique part of each of those would correspond to the Crispr RNA specific sequence, if you like, the rainbow colours that you referred to previously [from the CRISPRFinder figure], whereas the spacer region I would predict would be the path that binds to the tracrRNA.
MR FLYNN: Yes, so could you turn a couple of pages through in Professor Giffard’s first affidavit, to paragraph 77. That’s where he says, if you look at the second sentence, he says: “I understood there were critical property of individual Crispr RNA molecules”. So, he’s talking about the molecule, but he says: “Is that they each contain a transcribed spacer sequence”. Do you see that?
PROF THOMAS: Yes.
MR FLYNN: And what you understand him to be saying there is that an individual Crispr RNA molecule contains the RNA form of the spacer sequence in DNA. Do you agree with that?
PROF THOMAS: Well, it includes it. It includes it, but you can’t directly take the DNA sequence and just predict with certainty what the Crispr RNA is based on that sequence. There’s some processing involved.
MR FLYNN: Yes, but it will include what some or all of a transcribed spacer sequence. Correct?
PROF THOMAS: Yes, that’s correct.
MR FLYNN: And that’s why in the joint report you used the word, “encompass”, correct?
PROF THOMAS: Yes, I was trying to communicate that concept.
252 Professor Giffard’s next step was to find where the DNA sequence coding for tracrRNA is located in the bacterial genome. He said that he was aware from his own work prior to October 2012 that the stretches of DNA specifying the Cas proteins and the CRISPR array are in very close proximity. In support of this opinion he referred to Deltcheva (2011) which showed that in six out of seven species, DNA encoding the tracrRNA is located either immediately adjacent to or within the Type II CRISPR/Cas loci as defined by the presence of the Cas/Csn genes and the CRIPSR array.
253 Using that information, Professor Giffard searched for a DNA sequence that was either within or immediately adjacent to the S. thermophilus strain CRISPR/Cas locus that, once transcribed, produce an RNA molecule that is complementary to the repeat sequence of the CRISPR array. Put another way, he searched for a DNA sequence that was identical to the repeat sequence of the CRISPR array. He did this using a pairwise BLAST analysis where one of the pair is the entire S. thermophilus strain MN-ZLW-002 genome sequence (from NCBI) and the other is a single copy of the CRISPR repeat from that strain (from CRISRFinder). In this way, Professor Giffard was able to ascertain the location and sequence of the DNA coding for tracrRNA.
254 In the result, Professor Giffard was able to identify the DNA coding for both crRNA and tracrRNA and determine the endogenous crRNA and tracrRNA sequences for the Type II CRISPR/Cas system of the S. thermophilus strain MN-ZLW-002.
255 The respondents submitted that Professor Giffard’s identification of the DNA coding for the tracrRNA sequence was infected by an impermissible reliance on Deltcheva (2011) which provided him with the knowledge that tracrRNA is encoded by a gene separate from the CRISPR array but very close to or within the CRISPR-Cas locus. I agree with ToolGen’s submission that the respondents’ criticisms based on Professor Giffard’s reliance on Deltcheva (2011) ultimately falls away given my finding that it was likely that he had read Deltcheva (2011) and that it was common general knowledge of a microbiologist at the priority date.
256 The respondents also submitted that the CRISPRFinder was not common general knowledge of molecular biologists or microbiologists at the priority date. I accept, based on the evidence of Associate Professor Herold, that it was not common general knowledge of molecular biologists working in the field of gene editing in eukaryotic cells. However, I consider it was a tool that is likely to have been commonly used by microbiologists specialising in the study of CRISPR systems. In Giffard 1, Professor Giffard referred to CRISPRFinder as a publically available resource which he typically referred to in the course of his work. In his oral evidence, he acknowledged that it was likely he had not used the CRISPRFinder prior to, or in, October 2012 for the purpose of seeking to identify a CRISPR array. However, Professor Giffard’s evidence shows that CRISPRFinder was one of a number of tools available for that purpose and that it was the subject of a paper by Grissa et al (“Grissa 2007”) published in Nucleic Acid Research in 2007.
257 The fact that Professor Giffard may not have used the CRISPRFinder himself before October 2012 is not determinative of whether it was at that date common general knowledge in his field. In light of his evidence considered as a whole, including the description of the tool published by Grissa (2007), I find that CRISPRFinder was a well-known tool available for use by microbiologists engaged in CRISPR research in October 2012. On this issue, I consider it significant that the respondents did not directly challenge Professor Giffard’s evidence to that effect nor call evidence from any other microbiologist refuting it.
Identifying and characterising the mature crRNA and tracrRNA
258 The next step identified by ToolGen that the skilled addressee would need to take to perform the invention of the claims in other Type II CRISPR-Cas bacterial species was to identify and characterise the mature (fully processed) crRNA and tracrRNA.
259 ToolGen submitted that once the putative crRNA and putative tracrRNA sequences are ascertained in the manner previously outlined, the length and endpoints of the mature crRNA and tracrRNA could be ascertained by obtaining the bacterial isolate and using techniques which were standard as at October 2012.
260 In JER 2, Professor Thomas identified several techniques to identify the crRNA and tracrRNA molecules. In relation to identifying the crRNA molecule he stated:
To identify the crRNA molecule in its fully processed form, I would seek bacterial RNA expression data (RNAseq) from [sic] bacterial isolate in question. I would search that data for sequences corresponding to the putative crRNA sequence. This might provide me with the sequence of the processed/ functional form. If RNAseq data were unavailable, then I would seek to obtain the bacterial isolate for experimentation. If the bacterial isolate was available for experimentation, I would perform experiments such as northern blot/ RNAse protection and RNAseq to identify the length/ content of the crRNA.
261 Associate Professor Herold and Professor Giffard both broadly agreed with Professor Thomas’ approach, and Professor Giffard added that this approach was similar to the RNA sequencing (“RNA-Seq”) based approach taken by Deltcheva and her co-workers in Deltcheva (2011). RNA-Seq data provides what for present purposes may be described as a snapshot of the RNA in a cell at a given time.
262 In relation to identifying the tracrRNA molecule, Professor Thomas stated that he would follow the same approach as he took to identify the crRNA molecule. Associate Professor Herold and Professor Giffard both broadly agreed with Professor Thomas’ approach and Professor Giffard added that this approach was consistent with what he described in Giffard 2 with reference to Deltcheva (2011).
263 The authors of Deltcheva (2011) used RNA-Seq to characterise and quantify RNA derived from S. pyogenes strain SF-370. Professor Giffard stated that this involved: (1) extracting RNA from cells; (2) converting RNA to complementary DNA (cDNA); (3) sequencing the cDNA using high-throughput next generation sequencing technology and (4) using bioinfomatic techniques to map the sequencing data on the genome sequences of the organisms from which the RNA originated. Professor Giffard went on to state that during the preparation of purified RNA, half of the sample was treated with a compound (TEX) which depletes the RNA of processed transcripts, thereby enriching for primary transcripts. This enabled the authors to distinguish between RNA that had been produced in the cell by cleavage events (i.e processed RNA) and those that were the primary products of transcription (i.e. unprocessed RNA). This process yielded the nucleotide sequence including the beginning and end positions of any RNA identified and provided information on the relative abundance of particular RNA molecules, and whether the RNA was a product of a processing event within the cell. The authors identified both crRNA and tracrRNA.
264 In oral evidence, Professor Thomas ultimately agreed that if he was provided with the CRISPRFinder result for a particular bacterial species of interest coupled with the knowledge from a microbiologist that individual crRNA molecules each contain a transcribed spacer sequence, he could find sequences that correspond with the spacer sequences, and if he were confident that he could see a match to those sequences within the RNA-Seq Library, then he would have confidence that he had identified an endogenous RNA molecule that could be the crRNA.
265 Professor Thomas was questioned about whether high-throughput RNA sequencing technology was well-known before October 2012, and he gave the following evidence:
PROF THOMAS: It’s known, but reasonably specialised. It’s not the kind of thing, certainly, that [sic] – molecular biology labs generally would outsource that type of technology, because relatively complex machinery is required to perform the analysis. We – we were not doing RNA-Seq studies of that nature at the time. Some specialised labs were. You also need a bioinformatician to interpret the data, because you [sic] imagine you’re getting back an awful lot of information, so you need someone who’s experienced in the area to process that information and interpret it for you.
MR FLYNN: Yes. Now, if we read on in the joint report, you say if RNA-Seq data were unavailable, then you would seek to obtain the bacterial isolate for experimentation. Do you see that?
PROF THOMAS: Yes, I do, and the reason I – I’ve said that was that I think it’s actually probably pretty unlikely that RNA-Seq data would be available, because that’s an experiment that someone has to perform, so you would need to really have characterised that strain or that isolate in a lot of detail to inspire you to do an RNA Seq experiment. So actually, I suspect, given the nature of the searches we’ve been talking about, they could identify anything on the database that fits the criteria. The chances of that having an RNA-Seq data library available for that particular strain, I think, is probably pretty unlikely. But that’s – so that’s why I said it.
MR FLYNN: But you don’t know, as at October 2012, what was or wasn’t available in the library; is that right?
PROF THOMAS: No. No, I do not, but I just make the point that I think it’s unlikely.
266 The respondents submitted that Professor Thomas’ evidence establishes that it was unlikely that, for any particular strain of bacteria (leaving aside S. pyogenes) RNA-Seq data would have been available in October 2012. Professor Giffard agreed that in October 2012, he would have expected there to be no more than a small number of Type II repeat spacer expression data made available, and that, in 2013, there were only around six species other than S. pyogenes for which Type II CRISPR array expression data had been made available.
267 Associate Professor Herold gave evidence that the use of RNA-Seq data to identify the mature crRNA and tracrRNA components of a bacterial species other than S. pyogenes would require a tremendous amount of work including bioinformational analysis and experimental validation. Associate Professor Herold said:
… [T]he tracrRNA is required for specificity and also CRISPR RNA for the individual Cas proteins to work. So that would be the first step to identifying. The identification of the CRISPR is the first thing which would be difficult. You could do this bioinformatically but then you had to confirm experimentally, that would be the first step. And then identifying the tracrRNA, in October 2012 – at the time when the P1 came out and Jinek came out – the only tracr which has been described in the literature was S. pyogenes in the Deltcheva paper in 2011. And this was a Nature paper, again, showing this tremendous amount of work they required to get this up and running...So I would assume that it’s not as easy just taking a tracr and a CRISPR because we don’t even know where to find and where to look for and what size to be used. So it’s very, very difficult to get this working and would require very, very significant amount of work from a specialised lab.
(Errors in original).
Professor Thomas agreed.
268 One difficulty I have with this evidence is the use that Associate Professor Herold seeks to make of Deltcheva (2011). It may be accepted that Deltcheva (2011) was ground breaking even though, on the respondents’ case, Associate Professor Herold knew nothing of Deltcheva (2011), the focus of which was outside his field of expertise. Deltcheva (2011) was important, not so much because it provided the RNA-Seq for S. pyogenes, but because of its contribution to the understanding of the role of tracrRNA in directing the maturation of crRNAs. I give little weight to Associate Professor Herold’s evidence based on Deltcheva (2011).
269 The respondents also sought to support Associate Professor Herold’s views as to the difficulty involved in identifying mature tracrRNA in a bacterial species other than S. pyogenes by reference to Jinek. They submitted that Associate Professor Herold’s perception of the difficulties involved was confirmed by the fact that the authors of Jinek (who included the eminent microbiologists, Charpentier and Doudna) did not use experimentally validated tracrRNA from L. innocua and N. meningitidis. The supplementary figures of Jinek show that in evaluating the DNA cleavage of Cas9 orthologs related to S. pyogenes, the authors used predicted tracrRNA sequences based on the northern blot data published in Deltcheva (2011) for S. pyogenes.
270 The fact that the authors of Jinek relied on predicted tracrRNA sequences for L. innocua and N. meningitidis, was the basis for a suggestion in Associate Professor Herold’s evidence that, if the authors could have used the actual tracrRNA sequences derived from the bacteria using their own experimentally derived data, then they would have done so; the implication of them not doing so was, according to Associate Professor Herold, that it was too difficult even for them. However, as Associate Professor Herold acknowledged in his evidence, the authors do not suggest that identifying the tracrRNA for those bacteria would have been difficult.
271 In JER 2, Professor Thomas also identified northern hybridisation (or northern blot) and RNase protection as means to identify the mature crRNA and tracrRNA molecules. Professor Thomas accepted that these techniques were well-known in October 2012 and that both could be used to determine the length of RNA molecules. Professor Giffard also gave evidence that these techniques were well known at the priority date.
272 I find that both northern hybridisation (northern blot) and RNase protection were standard techniques used by the notional skilled addressee at the priority date, and that both techniques could be used to identify mature crRNA and tracRNA molecules.
273 In cross-examination, Senior Counsel for the respondents questioned Professor Giffard regarding potential difficulties involved in using these techniques each of which requires access to the bacterial isolate of interest. He gave the following evidence:
MR DIMITRIADIS: You would need to conduct some other sort of experimentation of the kind that you referred to, in order to – if you weren't using the RNA-Seq, is that right?
PROF GIFFARD: As I got fairly specific in my second affidavit, a combination of northern blots and primer extension, despite being old technology, is very effective and very robust and would be effective at identifying the ends of the molecules, you know, the exact natures of these molecules, and as a backup technique, the additional technique that was described by Deltcheva was the circularised RNA combined with PCR, which is essentially just a belt and braces thing; it’s just another armoury to make sure that you can really nail down exactly these molecules.
MR DIMITRIADIS: And each of those techniques that you’ve just referred to, require having a bacterial isolate in question, available, before experimentation, correct?
PROF GIFFARD: That is correct. Or I guess, in theory, someone else could have it and could send you the – send you a physical RNA preparation. That would not be a common thing but…in theory, if someone else extracted the RNA, then sent it to you, then you could experiment on that. But yes, you need to have an actual preparation of the RNA from the bacterial cell.
274 The evidence was quite vague as to how difficult it would be to obtain samples for use in a northern hybridisation or RNase protection experiment. Professor Thomas gave evidence that the difficulty with requiring a biological sample for experimentation was that it may not be possible to obtain if the DNA sequence of interest was not cultured in a laboratory.
275 Professor Giffard gave evidence that if the bacterial isolate was obtained, the experimentation process for northern hybridisation (northern blot) or RNase protection could then be conducted. He gave this evidence, which I accept, as to the difficultly involved in conducting such experiments:
MR DIMITRIADIS: Now, would you agree with this: if RNA sequence data or data that you are seeking to obtain from the experiments of the kind that you referred was available, that those – that work would be extensive and would require a significant amount of experimentation in order to obtain the data. Is that fair?
PROF GIFFARD: No. I don’t think it is because, as I mentioned, the dRNA-Seq in not essential to analyse the RNA molecules. And techniques such as northern hybridisation and primer extension are old, relatively low tech and even quite cheap experiments that can be carried out in a matter of days. And so, in theory, assuming everything goes right and with those – those methods, if you have got some reasonable skills in the lab, there’s no reason to think they wouldn’t go right. And it would be a – quite small project to use northern hybridisation and primer extension or RNase protection, or even the circularisation of the PCR. None of those are particularly challenging or particularly time consuming. I mean, I – as I have said, I had – I have published northern hybridisation, RNase protection and primer extension in 1993 and 1995, and I – part of that I did myself, and part of it my research assistants at the time did. And the northern hybridisation took a bit of time because we had a bit of trouble figuring out how to purify the RNA from oral bacteria, which are very robust cells that are hard to bust open, without breaking the RNA as well, but once we had the RNA it’s so similar to southern hybridisation, which I’ve been doing a lot. And it’s really within the remit of anyone with confidence in recombinant DNA technology and DNA detection technology to do these relatively straightforward experiments. In fact, they’re within the range of what could be done in undergraduate classes.
276 Professor Giffard’s evidence makes clear that northern hybridisation (northern blot) and RNAse protection do not require RNA-Seq to identify the mature crRNA and tracrRNA. Accordingly, the difficulties identified by Associate Professor Herold regarding the need for a bioinformatician to process and interpret the RNA-Seq data and experimental validation which requires next generation sequencing is not relevant to a consideration of these two alternate techniques. In my opinion, the evidence of Professor Thomas and Professor Giffard in relation to northern hybridisation (northern blot) and RNase protection techniques renders Associate Professor Herold’s evidence concerning the difficulties involved in using RNA-Seq largely irrelevant.
277 However, Professor Thomas identified two further difficulties regarding these techniques. First, the DNA sequence for the bacterial strain of interest must being publicly available. Second, for northern hybridisation (northern blot), the relevant RNA molecules (i.e. the putative crRNA and tracrRNA) must be identified before northern blot experiments can be carried out.
278 As to Professor Thomas’ first point on the public availability of the DNA sequence of interest, this would have been identified in the two preceding steps of, first, identifying a cas9 from another Type-II CRISPR-Cas9 system using the NCBI and, second, identifying the DNA coding for the tracrRNA using a pairwise BLAST analysis to compare a whole genome sequence (from NCBI) with a single copy of the CRISPR repeat from that strain obtained from CRISRFinder. As such, it can be assumed that by the stage that the skilled addressee attempts this step, they will have access to a publicly available copy of the DNA sequence of interest.
279 As to the identity of the crRNA molecule, Professor Thomas accepted in his oral evidence that this would be the spacer sequence of the CRISPR array or some portion of it. However, in earlier oral evidence and in Thomas 2, he explained that even with the DNA from the CRISPR array, it is difficult to identify the crRNA molecules because it is not possible to predict with certainty that the crRNA molecule is based on that sequence because of processing involved (from pre-crRNA to crRNA). However, he accepted that the crRNA would include some or all of a transcribed spacer sequence.
280 As to the identity of the tracrRNA molecule, Professor Thomas gave evidence that it could also be difficult to identify the tracrRNA because it would not be clear what part of the repeat sequence would be complementary to the tracrRNA and there is the potential for mismatches between the tracrRNA and the repeat sequence. In other words, even if the repeat sequence is known, it is not possible to know how much of it is going to be present in the tracrRNA and where any potential mismatches will be. Professor Giffard did not agree with Professor Thomas. He said that Professor Thomas’ argument about complementarity was not correct and disregarded the involvement of the tracrRNA in the pre-crRNA processing where the pre-crRNA itself is cleaved, which process is made possible by complementarity. On this issue, I prefer the evidence of Professor Thomas to that of Professor Giffard. I think Professor Giffard was most likely understanding the potential difficulties that may arise due to mismatches between the tracrRNA and the repeat sequence.
Identifying and validating the PAM sequence which the Cas9 of a non-S. pyogenes bacterial species recognises
281 The next step identified by ToolGen that the skilled addressee would need to take to perform the invention of the claims in other Type II CRISPR-Cas bacterial species was to identify and validate the PAM sequence which the Cas9 of that species recognises in eukaryotic cells. ToolGen relied on two approaches discussed in Associate Professor Firestein’s evidence: the first involved using a PAM variant library in vitro or in vivo and the second involved using an in silico approach (experimentation done by computer).
PAM variant library in an in vitro and/or in vivo system
282 In Firestein 1, Associate Professor Firestein set out three methodologies to use a PAM variant library and test cleavage in an in vitro and/or in vivo system.
283 Associate Professor Firestein set out his first methodology for identifying PAM variants for Cas9 orthologs homologous to S. pyogenes which included:
(a) Using high scale cloning methods to generate a PAM variant library which represents all possible PAM variants for S. pyogenes up to eight nucleotides in length.
(b) Constructing a sgRNA with the same sequence as the composition taken from S. pyogenes as shown in Figure 1A of P1 (“S. pyogenes sgRNA”) which would be customised by replacing the 20 base pair portion of the crRNA with a different sequence of interest.
(c) Performing a DNA cleavage assay such as that described in experiment 1 of P1, where the Cas9 is co-expressed with the sgRNA and is incubated with the PAM variant library.
(d) Isolating, purifying and deep sequencing (using next generation sequencing) the cut plasmid DNA generated from the cleavage assay to identify the different putative PAM sequences. A bioinformatician would be required to analyse the sequencing data.
284 Associate Professor Firestein stated that the second methodology involved the same steps as the first methodology, except he would specifically test the four cas9 orthologs identified in Jinek, namely, L. innocua, S. thermophilus, C. jejuni and N. meningitides. He said that he would test these homologous orthologs with the S. pyogenes sgRNA as he would expect that Cas9 orthologs, when highly homologous to each other, may exhibit similar activity even when interchanging certain system components (i.e. the sgRNA).
285 Associate Professor Firestein said that his third methodology involved the same steps as the first methodology, except instead of using the S. pyogenes sgRNA, he would construct either a sgRNA or a crRNA:tracrRNA duplex based on the endogenous crRNA and tracrRNA sequences for a particular bacterial species. I have previously discussed the process of identifying the endogenous crRNA and tracrRNA as explained in Professor Giffard’s evidence. Associate Professor Firestein said that he would prefer the approach of using a crRNA:tracrRNA duplex (by which I understood him to mean a crRNA and tracrRNA not fused into a single guide RNA), but that if he were to design and construct a sgRNA, he would use the approach taken in Jinek.
286 Use of a crRNA:tracrRNA duplex involves introducing the Cas9 and the mature crRNA:tracrRNA duplex to the pool of candidate plasmids and performing an in vitro cleavage assay as set out in P1. The cut plasmids are then isolated, purified and deep sequenced.
287 The in vivo PAM approach involves inserting the PAM variant library into a eukaryotic system so that every cell only has one PAM variant sequence. The cells express a RFP-GFP (red and green fluorescent protein) reporter system that allows for screening to identify GFP (green fluorescent protein) positive cells that have undergone CRISPR-Cas9 cleavage using a barcode approach. The GFP positive cells would then be isolated by flow cytometry and specific DNA fragments from the GFP positive cells as well as GFP negative cells would undergo next-generation sequencing to identify the PAM variants that are enriched in the GFP-positive cells and depleted in the GFP negative cells. Associate Professor Firestein gave evidence that since this is occurring in eukaryotic cells, a “barcode” would be externally placed as part of this reporter which would enable the skilled person to map back what the sequence of that PAM variant is in cases where cellular repair mechanisms may cause insertion or deletion across the PAM site itself.
288 The respondents submitted that because Associate Professor Firestein had never used a PAM variant library before or undertaken the computational analysis required as part of the analysis (which would require the expertise of a bioinformatician) it could not be said that this was routine work. Additionally, Associate Professor Herold gave evidence that at the priority date, although he had some familiarity with the approach taken by Associate Professor Firestein, it was not routine. In relation to the creation of a PAM variation library. Associate Professor Herold said:
…I would not say that is something in particular for me or a general normal molecular biological lab to do that as a routine work and could cause significant amount of work, and the chances of success would, in my opinion, be relatively low.
289 He identified several other issues with the use of a PAM variant library in an in vitro or in vivo system.
290 In Herold 2, Associate Professor said that he did not think the in vitro PAM variant library approach would be successful unless a large amount of time and effort was invested. He said:
[37] I do not consider that A/P Firestein's in vitro PAM approach would be successful in identifying other PAM sites recognised by a Cas9 in eukaryotic cells, unless a very large amount of time and energy was invested in the process. The approach involves the generation of 65,536 unique plasmids, representing all possible candidate PAM sites in an eight base pair sequence (4*4*4*4*4*4*4*4), of which only a very low number of plasmids would be cleaved…
[38] For example, if A/P Firestein's in vitro PAM approach was used for S pyogenes, a maximum of eight out of 65,536 plasmids (i.e. 0.01 % of all candidates generated) would be cleaved (the eight plasmids represent all four possible "NGG" variations, and all four possible "NAG" variations, in the PAM). Each cleaved plasmid would represent only one out of 65,536 plasmids (i.e. 0.0015% of all plasmids generated).
291 In oral evidence, Associate Professor Herold corrected his calculations by a factor of 1,000 and said that 8,192 out of 65,536 plasmids (12.5%) would be cleaved. Ultimately, he also accepted that the detection was no longer “almost impossible” but a “possibility” with no guarantee that any plasmid would be cleaved. When asked by me whether the probabilities of success were remote or something more, he suggested they would be something more.
292 Associate Professor Firestein gave evidence that even at low percentages, deep sequencing technology is sensitive enough to detect cut plasmids at low levels and Associate Professor Herold ultimately agreed with this evidence. The respondents had several issues with Associate Professor Firestein’s reliance on deep sequencing technology which, he accepted, would require the use of a next generation sequencer. Associate Professor Herold gave evidence that although next generation sequencers were available at the priority date, they were not standard equipment. Associate Professor Firestein accepted this and said that next generation sequencers would have only been available at a commercial laboratory for a fee or at core facilities in academic laboratories. Associate Professor Herold agreed with this.
293 Associate Professor Herold also gave evidence that commercially-sourced assays containing unique plasmids are generated in a Guassian distribution with concentrations of some candidate plasmids being very low, resulting in an uneven distribution of variants in the PAM library. Thus, if the correct PAM sequences are in low concentrations, it would be even more difficult to identify these PAMs using Associate Professor Firestein’s methodology.
294 Associate Professor Firestein accepted the premise of Associate Professor Herold’s evidence, but made clear that he did not accept the cut plasmid would not be detected and that the concerns raised by Associate Professor Herold would not arise in practice unless all of the 8,000 PAM variants respond to an active PAM that is present at low frequencies. Additionally, he suggested that this issue could be overcome by either conducting the assay so that each variant is represented in hundreds or thousands of plasmid copies or through quality control checks, whereby libraries that do not have appropriate representations of different variants that are sought would be discarded. Associate Professor Herold agreed that you could increase the number of molecules but that one may still struggle to find cut plasmids using next generation sequencing. According to Associate Professor Herold, use of next generation sequencing would significantly increase costs and take three to six months. Associate Professor Firestein disagreed and stated that because each molecule with a specific PAM would be represented 1000-fold times, it could easily be identified by next generation sequencing.
295 Associate Professor Herold identified one further issue pertaining to Associate Professor Firestein’s first and second methodologies which use the S. pyogenes sgRNA. His evidence was that Associate Professor Firestein’s first and second methodologies, which used a 20 nucleotide target sequence and an eight nucleotide candidate PAM site, would limit the detection of PAMs for a particular species where the target sequence and PAM site have nucleotides different to 20 and eight respectively. In their closing submissions, the respondents rely on two examples which became known after the priority date. These are a C. jejuni Cas9 which recognises a 22 base pair target site and a seven nucleotide PAM (NNNNACA), and N. meningitidis Cas9 which recognises a 24 nucleotide target sequence and an eight nucleotide PAM (NNNNGATT).
296 Whilst I accept that using a S. pyogenes sgRNA which is limited to 20 variable nucleotides for the target sequence and an eight nucleotide PAM may result in some PAM sequences not being identified, in circumstances where Associate Professor Firestein has proposed as an alternate approach the use of a duplex, which avoids this complication, and where he has specifically stated that he prefers this approach to using a sgRNA, I find that the issues identified in relation to the use of a sgRNA could not constitute a significant problem for him. But it does not seem to me to be likely that the notional skilled addressee working with P1 would adopt that approach given that it teaches the use of a sgRNA rather than a crRNA:tracrRNA duplex.
297 Associate Professor Herold identified two further issues that relate only to Associate Professor Firestein’s in vivo approach. As mentioned above, the in vivo approach relies on using eukaryotic cells that express a RFP-GFP reporter system that allows for screening to identify GFP positive cells that have undergone CRISPR-Cas9 cleavage using a barcode approach.
298 Associate Professor Herold referred to the cellular repair mechanism (“NHEJ”) in eukaryotic cells that repairs breaks in DNA. He said that a cut made at the PAM site may have been repaired and mutated by NHEJ which may make it impossible to determine whether the plasmid DNA was cut. In response, Associate Professor Firestein stated that he would address this issue by generating an alternative pooled PAM variant library that incorporates a barcode DNA sequence upstream of the target PAM site, which would enable the identification of a PAM variant even where the PAM sequence may have been lost due to insertions or deletions caused by NHEJ. Associate Professor Herold said that this approach would require much more additional work and there would be no guarantee that the barcodes were not destroyed by the Cas9 or cellular machinery. Associate Professor Firestein explained that he did not see the possibility that the barcodes may be destroyed as an issue as there were usually only really small deletions of a few nucleotides and if they were to occur this would only be in some proportion of the approximately 1,000 copies of the particular variant available for analysis.
299 Associate Professor Herold said that to use barcodes and the RFP/GFP reporter in the cloning step of the in vivo approach would require that the task be outsourced to a commercial laboratory which would cost a considerable amount of money. I accept that evidence.
300 Professor Thomas also said that Associate Professor Firestein’s in vitro or in vivo approaches of finding the PAM for a particular Cas9 created a so called “catch 22” issue, in that it is necessary to have a functional guide RNA in order to cleave the plasmid library and the investigator cannot know whether they do without first knowing the PAM for the Cas9. Associate Professor Firestein said that this is only an issue where the in vitro or in vivo DNA cleavage assay produces a negative result. Professor Thomas agreed and stated that the issue was that if there was a negative result, you could not know whether that was because there was no PAM or because the guide RNA and Cas9 was not complexing properly to cut if a PAM was present.
301 The respondents submitted that the catch 22 arises regardless of whether duplex guide RNA or a sgRNA is used. Senior Counsel for ToolGen asked Professor Thomas and Associate Professor Herold whether this issue could be overcome by adopting Associate Professor Firestein’s third approach of using the mature crRNA and tracrRNA for a specific species to prepare a crRNA:tracrRNA duplex to identify species-specific CRISPR Cas9 PAM sites. He suggested that this would overcome one unknown variable in the “catch 22” issue which is whether or not the sgRNA complexed with the Cas9 is able to cut as no sgRNA is used. Associate Professor Herold responded that this removes the issue of something going wrong in the fusing process to create a sgRNA but the issue is still whether the right mature crRNA and tracrRNA are identified. I accept that evidence. However, I also note that the identification of the mature crRNA and tracrRNA molecules could be identified using either the RNase protection or northern hybridisation (northern blot) approaches proposed by Professor Thomas.
302 In Firestein 1, Associate Professor Firestein explained his alternate approach of in silico identification, which uses in silico analysis of the CRISPR/Cas9 gene locus for each bacterial species to identify the putative PAMs for Cas9 orthologs. He set out the following methodology:
(a) Search each particular bacterial species genome for the presence of a CRISPR array, which, based on Figure S1 of Jinek, he would expect would be in the vicinity of the Cas9 gene. Identify within the CRIPSR array the repeat and spacer sequences and extract the spacer sequences.
(b) Identify the bacterial phage DNA target site (protospacer) from which the spacer originated by performing an NCBI BLAST search using the DNA of the spacers identified in subparagraph (a) above. The identified bacterial phage sequences that are identical or highly homologous (greater than 90%) to the sequences of the spacers would be designated as putative target DNA sites.
(c) Undertake computational analysis of the target DNA sites identified in subparagraph (b) above to identify a consensus sequence that defines the PAM site.
(d) Verify experimentally the putative PAMs identified by performing an in vitro cleavage assay as set out in experiment 1 of P1.
303 In Firestein 1, Associate Professor Firestein gave this evidence concerning the in silico approach:
[261] This in silica approach could be undertaken using simple computational tools (or undertaken manually). Where the genomic sequence data from the invading bacterial phage is available, this analysis can be done in a very short period of time (hours). The statistical power of this analysis is proportional to the number of spacers and protospacers that can be analysed and used to build a consensus plot for each Cas9 ortholog.
304 Associate Professor Herold agreed that if it is a “consistent PAM site” the in silico approach would identify the PAM. There was no evidence of any “inconsistent PAM site” identified by the respondents. Additionally, both Associate Professor Herold and Professor Thomas agreed that the in silico approach eliminates many of the issues raised in relation to the pooled variant library approach. However, they both raised several issues in relation to this approach.
305 Professor Thomas and Associate Professor Herold gave evidence that the phage sequence containing the PAM will not necessarily have been available on the NCBI database at the priority date. However, both accepted that as at the priority date, this was knowledge that would have been relevantly held by a microbiologist and not a molecular biologist. Professor Giffard did not give any evidence concerning the availability of phage sequences at the priority date. In those circumstances, the evidence does not establish that the in silico approach is feasible at least to the extent that it depended on the availability of phage sequences that were identical or highly homologous to the relevant spacer sequences.
306 Associate Professor Herold also made the point that it cannot be assumed that the phage sequences in the NCBI database are complete, correctly entered and correctly annotated. However, he accepted that he had no personal knowledge as to how extensive the phage, plasmid or genomic data was on the NCBI database at the priority date, and did not provide any specific examples where an incorrect, incomplete or untagged phage sequence would have prevented PAM identification.
307 In oral evidence, Professor Giffard stated that errors, truncated sequences and incomplete sequences in the NCBI database are rare. He viewed the likelihood that any sequence would be completely incorrect to be “probably zero” and stated that where there is a truncated or incomplete sequence, these still could include the information sought.
308 ToolGen rely on two ways to overcome the difficulties identified by Professor Thomas and Associate Professor Herold. First, ToolGen submitted that neither Professor Thomas nor Associate Professor Herold were able to provide any specific examples where an unavailable phage sequence would have prevented PAM identification. Second, ToolGen submitted that Professor Thomas readily accepted, to the extent that some genomic data was not present in the database, its absence was readily addressed by authors of Fonfara (2013) by searching for other isolates of the same species. Fonfara (2013) was published online in November 2013 and is a post priority date paper. I accept that this could be done. But this still assumes that the relevant phage sequence would be available to the investigator.
309 Associate Professor Herold also raised several other issues, including:
(a) The information in Jinek identifying the position and arrangement of individual components of the CRISPR/Cas9 locus is not generally applicable to all CRISPR/Cas 9 loci and varies between different bacterial species and strains.
(b) There is also a possibility that multiple CRISPR arrays exist in a single bacteria which would create difficultly.
(c) There is the further possibility that the fragments of DNA that make up the spacer may no longer be identical to the target site because of mutations.
310 I accept what appears in subparagraph (a) which is confirmed by the evidence more generally. I also accept that the other difficulties identified by him may arise.
Designing and constructing the sgRNA
311 ToolGen submitted that once it is appreciated that the disclosure of the sgRNA in P1 is not limited to the specific form of sgRNA used in the experiments of P1, the skilled addressee or team would use Jinek to design and construct the sgRNA. Accordingly, ToolGen’s submission on constructing a sgRNA from a bacterial species other than S. pyogenes relies on Jinek either being part of the disclosure of P1 by incorporation, or part of the common general knowledge of a microbiologist and molecular biologist. As I have previously found that Jinek does not form part of the disclosure of P1, and was not common general knowledge of a microbiologist or molecular biologist at the priority date, the following considerations are only relevant in circumstances where either of those conclusions is wrong.
312 In JER 1, Associate Professors Firestein and Herold and Professor Thomas agreed that the reference to Jinek in P1 “is a reference to the data in Jinek describing a technical advance regarding the fusion of tracrRNA and CRISPR RNA to generate a single guide (single chain chimeric) RNA.” Further, all three experts agreed that the tracrRNA and crRNA could be fused together and tested in a CRISPR/Cas9 cleavage assay in vitro using the principles detailed in Figure 5 of Jinek, which states that the chimeric RNA is generated by fusing the 3’ end of the crRNA to the 5’ end of the tracrRNA.
313 In Jinek, the authors identified the essential portions of tracrRNA and crRNA of an S. pyogenes sgRNA capable of guiding Cas9 mediated DNA cleavage. In oral evidence, Professor Thomas agreed that while one would not simply supplant the minimal portions of crRNA and tracrRNA for the S. pyogenes sgRNA shown in Jinek and apply this to a non-S. pyogenes Cas9 system, the minimal portions of crRNA and tracrRNA for a non-S. pyogenes sgRNA could be determined using a “classic experimental approach”. The “classic experimental approach” was described as starting with the full length of the mature crRNA and tracrRNA molecules and deleting regions of the molecules until one can determine what is the minimal amount required for Cas9 mediated cleavage.
314 Professor Thomas also gave evidence that the experiments undertaken to identify the minimal portions of crRNA and tracrRNA in Jinek adopt this same classic experimental approach. Associate Professors Firestein and Herold agreed that this approach could be employed to determine the minimal amount of crRNA and tracrRNA for any other Cas9 species.
315 Jinek also makes use of a radiolabeled assay and radioactive gel to test whether or not certain truncated forms of both crRNA and tracrRNA and the Cas 9 will cleave double-stranded DNA in vitro. The experts agreed that a cleavage test would be required to confirm that the truncated portions were indeed the essential portions of crRNA and tracrRNA required for Cas9 cleavage. When asked by Senior Counsel for ToolGen how difficult it would be to perform this radiolabeled assay, Professor Thomas said that it would involve a significant amount of work because of the nature of the assay and that it uses radioactively labelled oligonucleotides as the type of DNA sequence. Associate Professor Herold stated that it was not something standard that he would have done at the priority date and agreed with Professor Thomas that it would require a significant amount of work which could take up to three months or more. Associate Professor Firestein said that he did not have experience with radiolabeled assays and so would use the standard cutting assay shown in Figure 1B of P1. Associate Professor Herold agreed this approach could be taken but that he would be cautious about adopting it because it is much less sensitive than the radiolabeled assay.
316 Associate Professor Herold gave evidence that to construct a sgRNA from the endogenous crRNA and tracrRNA components of other Cas9 species of interest that had not yet been described would be very difficult and would require a significant amount of work from a specialised laboratory. The difficulties with identifying and characterising the mature crRNA and tracrRNA molecules have already been discussed above in a preceding step.
317 Associate Professor Herold raised two further issues with starting with the full length of the mature crRNA and tracrRNA molecules. First, he stated he would not have done this because at the priority date a sgRNA using a full length tracrRNA had not been shown to work. Second, he stated that he would have concerns about using a full length tracrRNA due to the possibility of a more complex RNA structure causing an interferon response which cause cell death in a eukaryotic cells. However, ToolGen do not simply propose using the full length tracrRNA and crRNA molecules and stopping there. What it proposed is to replicate the approach taken by the authors of Jinek and use the mature crRNA and tracrRNA molecules as a starting point from which to cut down portions to identify the essential portions of crRNA and tracrRNA needed for Cas9 mediated cleavage.
318 Against that complex background it is now necessary to ask whether the invention of claims 1 and 10 of the patent application are sufficiently enabled by the disclosure of P1. In particular, did P1, as at the priority date, provide sufficient information to the notional skilled addressee armed with the common general knowledge to perform the invention of the claim without undue burden over the whole scope of each claim? In deciding this question it is of course necessary to take account of my previous findings as to who is the notional skilled person or skilled team. It is also necessary to have regard to the nature of the invention of the claims, the field of technology, and the breadth of the relevant claims.
319 As to the nature of the invention, I do not think P1 discloses any principle of general application. It does not represent that all, or substantially all, system components (including a particular Cas9) derived from other bacterial species that have a Type II CRISPR/Cas system can be used to cleave DNA in eukaryotic cells, and does not state or imply that there is any reasonable scientific basis for concluding that most, or a substantial number of the many different bacterial species with a Type II CRISPR/Cas system, would be suitable for use in the compositions or the methods of the claims.
320 P1 shows that the inventors tested components derived from S. pyogenes only. There is nothing said in P1 which would indicate that S. pyogenes was likely to be representative of other bacterial species with a Type II CRISPR/Cas system or that the results of the experimentation with S. pyogenes derived components provided any reasonable scientific basis for inferring that Cas9 polypeptides derived from other bacterial species could also be expected to cleave DNA in eukaryotic cells.
321 Those observations are relevant to enablement because, as will be apparent from what I have previously said, P1 does not provide the skilled addressee with any encouragement, whether by the elucidation of a general principle or a series of working examples, that a system using Cas9 components derived from other bacterial species can reasonably be expected to cleave DNA in eukaryotic cells.
322 If, as I have found, P1 is not directed to a skilled team including a microbiologist, it follows that the claims are not sufficiently enabled. The molecular biologist is not told by P1 what other bacterial species have Type II CRISPR/Cas systems or how to determine the endogenous crRNA and tracrRNA sequences for such a species. The papers relied on by Professor Giffard when seeking to ascertain those sequences for S. thermophilus were not the common general knowledge of the molecular biologist and the CRISPRFinder, although a standard tool used by microbiologists, was not a standard tool used by molecular biologists. Nor was Professor Giffard’s knowledge as to the proximity of the Cas1, Cas2, Cas9 and the CRISPR array to each other in the bacterial genome as a means of identifying bacteria with a Type II CRISPR/Cas system common general knowledge of molecular biologists. Further, for the reasons previously stated, I do not accept that deficiencies in the common general knowledge of molecular biologists could be filled by the use of literature searches that might generate results that included Bhaya (2011), Makarova (2011) or Deltcheva (2011). Accordingly, the following considerations are only relevant in circumstances where I am wrong about whether a microbiologist is a skilled addressee for P1 and the skilled team has the benefit of the skills and common general knowledge of a microbiologist such as Professor Giffard.
323 In his written evidence, Associate Professor Firestein appears to have accepted that not all CRISPR/Cas9 systems that work in vitro or in prokaryotic cells are necessarily able to cleave target DNA in eukaryotic cells. In Firestein 2, Associate Professor Firestein stated at para 129:
Furthermore, although AP Herold states that he is now aware that not all CRISPR/Cas9 systems that work in vitro or in prokaryotic cells are “necessarily able to target and cleave DNA in eukaryotic cells”, in my opinion, this statement acknowledges that some systems that work in vitro also work in vivo. In this regard, my approach does not assume that in vitro activity correlates with in vivo activity, but rather tests and identifies systems that show activity in both contexts. Indeed, there are a number of reports in the literature that demonstrate multiple CRISPR/Cas9 systems, derived from different bacterial species, which are able to target and cleave genomic DNA in eukaryotic cells (as I discuss below in relation to Hou et al (N. meningitidis), Ran 2015 (S. aureus), Muller et al (S. thermophilus), Kim 2017 (C. jejuni) and Chatterjee et al (S. canis)).
324 It is clear from the context in which these observations were made that Associate Professor Firestein was referring to Type II CRISPR/Cas9 systems. I note that each of the articles to which he refers is concerned with a Type II CRISPR/Cas9 system derived from different bacterial species.
325 In my opinion, it would have been apparent to the skilled team as at the priority date that there was considerable uncertainty as to whether or not a CRISPR/Cas9 system derived from any particular bacterial species other than S. pyogenes would work in eukaryotic cells. The experts agreed that the ability of a CRISPR/Cas9 system to work in eukaryotic cells cannot be assumed by activity shown in an in vitro DNA cleavage assay and that additional work is required to test the CRISPR/Cas9 system in eukaryotic cells. They also agreed that the in vitro DNA cleavage assay is a relatively straightforward first approach that they would have undertaken to test a CRISPR/Cas9 system and, if it showed cleavage activity, would lead them to test the system in a eukaryotic cell.
326 Both Professor Thomas and Associate Professor Herold said that they would cease work on the candidate if it failed to show activity in vitro, although Associate Professor Firestein said that he would not assume that a candidate that did not work in vitro would not work in vivo. However, he did not refer to any examples in which a system that worked in vivo would not work in vitro. I regard Associate Professor Herold’s and Professor Thomas’ approach as more representative of the thinking of the notional skilled team on this topic.
327 Associate Professor Herold and Professor Thomas were also of the view that significant experimental work would need to be done to validate the use of the system in eukaryotic cells. Professor Thomas gave evidence that this would require conducting in vivo experiments at a wide range of different target sites and in a wide range of different cell lines. Associate Professor Firestein said in JER 1 that those steps would be straightforward and that the time taken to perform the work could depend on how much optimisation would be required for any particular system. The evidence of all three witnesses was somewhat vague as to how long such work would take but I am persuaded that it would involve a multi-step process requiring a significant amount of work with each step in the process dependant on the success of the previous step.
328 The skilled team would not know whether its work was likely to yield a product capable of achieving double stranded breaks at target locations in eukaryotic cells until the relevant candidate had been trialled and validated by in vitro and then in vivo experiments.
329 In relation to Associate Professor Firestein’s statements in JER 1 and his affidavit evidence suggesting that experimental validation of a Type II CRISPR/Cas9 system derived from other species would be straightforward, I refer to the following exchanges in the concurrent evidence:
MR DIMITRIADIS: Would you agree, Professor Firestein, that your approaches that you’ve discussed in your affidavit would involve a very significant amount of work – a research project – firstly?
ASSOC PROF FIRESTEIN: It’s difficult for me to – to speculate on that. It may involve a lot of work or it may be done quite quickly because, you know, if I bring up the case of streptococcal canis again, this is a system that’s nearly 90 per cent homologous to strep pyogenes. It works with the same – well, not the same but let’s say the tracrRNA is exactly the same, the crRNA differs by only one nucleotide.
Using that type of system, it could be repurposed quite quickly. If I had to go through hundreds of different CRISPR Cas9 systems, then, obviously that would be a significant amount of work. But it may also be that I – I hit upon a system much more quickly than that. And I think the important thing is that this is – that’s why his approach and the same approach applies for every CRISPR Cas9 system that I’m utilising.
MR DIMITRIADIS: Professor Firestein, you used the language “I can’t speculate”. I suggest to you that that is because you have never engaged a process of this kind seeking to identify a component of a CRISPR Cas9 system from another bacterial species, correct?
ASSOC PROF FIRESTEIN: It is true that I have never sought to identify a CRISPR Cas9 from another species. And I refer back to my comments on that, that I’ve not found the NGG to be a practical limitation. The reason that I cannot speculate is, based on my broader experience in research, that sometimes we set out a logical approach to [sic] certain problem, we conduct experiments, sometimes those experiments work quite quickly, sometimes they take longer. And I also would, in that respect, disagree with Professor Herold’s assertion that a Nature paper necessarily implies that there has been a lot of work involved.
330 Associate Professor Firestein therefore accepted that at least in some cases the amount of work involved in using a system derived from another bacterial species would be significant, but that in other cases the work could be done quite quickly. He cites as an example a system derived from S. canis that he suggested could be developed quite quickly, because it is nearly 90% homologous to S. pyogenes. His evidence was, in effect, that if he “hit upon” a system like that, it could be developed very quickly. There are several points to make about this evidence.
331 The evidence suggests that the system derived from S. canis was first characterised in a paper authored by Chatterjee et al published in 2018 (“Chatterjee 2018”) which described a Cas9 protein derived from S. canis and the PAM sequences it recognised. I will say more about this paper shortly, but it constitutes post priority date information that would not have been available to the skilled team. In particular, Associate Professor Firestein’s reference to the S. canis system being nearly 90% homologous to the S. pyogenes system seems to be derived from Chatterjee (2018) which reported S. canis Cas9 as having 89.2% sequence similarity to S. pyogenes Cas9. The sequence appears not to have been characterised or functionally validated until Chatterjee et al undertook that work.
332 However, I accept that some support for Associate Professor Firestein’s reasoning is found in Jinek. This paper showed that of four S. pyogenes orthologs investigated in vitro, two Cas9 derived from L. innocua and S. thermophilus which are bacterial species that are homologous to S. pyogenes (54% and 58% homology respectively) could be complexed with the same S. pyogenes sgRNA to cleave DNA in prokaryotic cells. This was not so in the case of two Cas9 derived from C. jejuni and N. meningitidis which are bacterial species that have lower homology to S. pyogenes Cas 9 (16% homology for both). I also note that Associate Professor Herold and Professor Thomas agreed that homology between S. canis and S. pyogenes Cas9 and their crRNA and tracrRNA are nearly identical and that it is therefore very likely that the Cas9 and sgRNA components of these systems could be interchanged to mediate CRISPR/Cas9 cutting. Therefore, whilst I do accept the proposition that it may be easier to develop a functional system in bacterial species that are highly homologous to S. pyogenes, including because the same S. pyogenes sgRNA or a very similar sgRNA can be used, I think it must also follow that there is likely to be a very substantial proportion of Cas9 derived from other bacterial species that are not sufficiently homologous to S. pyogenes Cas9 to enable use of the same sgRNA or a very similar sgRNA to cleave DNA in eukaryotic cells.
333 Importantly, claims 1 and 10 are not confined to Streptococcus Cas9 polypeptides and extend to any bacterial species with a Type II CRISPR/Cas system whether or not within the Streptococcus genus and whether or not the Cas9 protein for that species is highly homologous to Cas9 derived from S. pyogenes. The disclosure required of P1 is that it be such as would enable the skilled team armed with the common general knowledge to make all, or substantially, all, embodiments within the scope of the claims without undue burden. Associate Professor Firestein’s evidence concerning the ease with which the sgRNA used in Jinek could be re-purposed to Cas9 from other species would not hold true across the scope of the claims.
334 The respondents relied on the paper by Ran et al published in Nature in 2015 by a group of researchers associated with the Broad Institute which appears to be a collaboration involving MIT and Harvard. In their closing submission, ToolGen objected to the respondent’s reliance on Ran (2015) and submitted that it and other post priority date papers should be given no weight on the issue of undue burden for the following reasons:
(a) no persons who did that work were called to give evidence;
(b) the articles are notable for their lack of reporting any difficulty at all, or any difficulty which was not easily overcome;
(c) the reviewers of the article were not called to give evidence to say precisely what aspect of the work they regarded as qualifying it for publication;
(d) the articles often involved many different tasks and experiments, and it is impossible to ascertain how much work was involved in any individual task relevant to the specific questions of enablement in this case;
(e) it is impossible to separate the work which may have gone into writing up and reporting upon the experiment in a form suitable for publication in a journal from the actual scientific work involved;
(f) the articles were not contemporaneous records of the amount of work done, but rather were highly polished accounts of work done, specifically drafted for the purpose of impressing reviewers and achieving publication; and
(g) the respondents have adduced no evidence that any of the work the subject of those papers was inventive or non-routine, and mere publication does not prove it to be so.
335 Each of the articles to which ToolGen’s submission was directed was admitted into evidence without objection or limitation as to the use which might be made of it. That said, in deciding what weight to give Ran (2015) and other post priority date papers I have had regard to ToolGen’s submission. However, I have also had regard to the fact that at least some of these papers were relied on by Associate Professor Firestein as demonstrating that multiple CRISPR/Cas9 systems derived from different bacterial species are able to target and cleave genomic DNA in eukaryotic cells. He accepted, speaking in the context of Ran (2015), that a paper published in Nature would be one that the publisher and reviewers considered to be a fairly substantial piece of original research or, in his words, “substantial in its concept and advancement of the field”. He did not accept that you could draw any inference as to the amount of time the research work took. I agree with that, but I also accept Professor Thomas’ evidence who, also speaking of Ran (2015), said that it must have reflected a very large amount of work.
336 Both Associate Professor Herold and Associate Professor Firestein referred to Chatterjee (2018) which was submitted in May 2018 and published in October 2018 in Science Advances as a research article. Chatterjee (2018) states that while numerous Cas9 homologs have been sequenced, only a handful of Streptococcus orthologs have been characterised or functionally validated. The authors describe how they characterised an orthologous Cas9 protein from S. canis which, as mentioned earlier, had a sequence homology of 89.2% similarity to that of S. pyogenes Cas9. They explain in some detail how they determined the PAM sequences recognised by S. canis Cas9 first in vitro and then in vivo in human cells.
337 Another paper by Kim (2017) submitted in October 2016 and published in February 2017 in Nature Communications (whose authors included Jin-Soo Kim and other researchers that are named inventors of P1 and the patent application) noted that several CRISPR/Cas9 orthologs had been used for genome editing. Kim (2017) describes a new Cas9 ortholog derived from C. jejuni an advantage of which is said to be its smaller size. The paper describes the steps taken by the authors to determine the PAM sequence for this Cas9 ortholog and to optimise the length of the sgRNA before delivering the system via an adeno-associated virus (AAV) to mammalian cells for in vivo genome editing.
338 With reference to Ran (2015), the respondents submitted:
179. The nature and standard of work required to develop another Type II CRISPR/Cas9 system for use in gene editing in eukaryotic cells are well illustrated by reference to the Ran paper. This was a publication in Nature, a prestigious journal, where the authors studied six CRISPR/Cas9 systems which showed cleavage in vitro but only identified two species which showed cleavage in vivo (including S. Aureus).
…
180. Ran was by members of the Broad Institute, which was not a typical academic laboratory, because of its breadth of expertise. The paper represents an enormous body of work, and is a combination of many peoples’ efforts. Its publication in Nature reflects the cutting edge nature of the research involved. It was an original piece of research that is substantial in its concept and advancement of the field.
339 The research work the subject of Ran (2015) involved more than finding and validating another bacterial species that could be used in place of S. pyogenes. It appears that the researchers specifically focused on Cas9 derived from other bacterial species with a lower molecular weight than S. pyogenes Cas9 because these were thought to be better suited for delivery using AAV vectors. However, in my opinion, Ran (2015) does provide some insight into the work involved in identifying S. aureus derived Cas9, the crRNA and tracrRNA associated with it and, in particular, whether the work involved would constitute an undue burden.
340 Ran (2015) describes the steps taken by the researchers to determine the endogenous crRNA and tracrRNA sequences for Cas9 derived from the six species that were investigated, the PAM sequences for each of them (using a plasmid library) and the ability of each Cas9 paired with a single guide RNA to cleave target sites first, in vitro, and then in vivo in mammalian cells. This ultimately led the researchers to identify S. aureus Cas9 as one which produced “indels” (ie. insertions or deletions) with efficiencies comparable to S. pyogenes Cas9.
341 The authors of Ran (2015) state at page 186:
In search of smaller Cas9 enzymes for efficient in vivo delivery by AAV, we have previously described a short Cas9 from the CRISPRI locus of Streptococcus thermophiles LMD-9 (St1 Cas9, ~3.3 kb) as well as a rationally-designed truncated form of SpCas9 (ref. 18) for genome editing in human cells. However, both systems have important practical drawbacks: the former requires a complex protospacer-associated motif (PAM) sequence (NNAGAAW), which restricts the range of accessible targets, whereas the latter exhibits reduced activity. Given the substantial diversity of CRISPR-Cas systems present in sequenced microbial genomes, we therefore sought to interrogate and discover additional Cas9 enzymes that are small, efficient and broadly targeting.
(footnotes omitted)
In support of that statement the authors referenced Chylinski (2014) (co-authors including Makarova and Charpentier) published in 2014 and by Chylinski (2013) (co-authors including Charpentier) published in 2013 (both post priority date).
342 The authors of Ran (2015) then referred to their analysis of over 600 Cas9 orthologs with protein sizes approximately 1,350 and 1,000 amino acid residues in length. From the 600 Cas9 orthologs, the authors selected six candidates for profiling which involved ascertaining the crRNA and the tracrRNA for each Cas9, and designing a single guide RNA (sgRNA) for each of the six orthologs. The authors then identified the PAM sequence for each Cas9 by constructing a library of plasma DNA and performing an in vitro cleavage assay. The authors reported that the Cas9 orthologs, in combination with the sgRNA, successfully cleaved their targets in vitro. They then proceeded to investigate whether they would do the same in mammalian cells, after noting that “DNA cleavage activity in cell-free assays does not necessarily predict activity in mammalian cells”. Of the six orthologs tested, only S. aureus produced indels with efficiencies comparable to those of S. pyogenes. The authors’ investigation thereafter focused on S. aureus. It is apparent that by that stage of their work, they had successfully cleaved DNA in mammalian cells using components derived from S. aureus which they then sought to optimise in various ways.
343 I think Ran (2015) is useful in so far as it describes the work that its authors undertook in order to identify other Cas9 orthologs which could be used to cleave DNA in in vitro assays, and the work its authors took to confirm that S. aureus Cas9 could be used to cleave DNA in vivo in human cells. Ran (2015) and the various other post priority date papers also tend to show that for some years after the priority date there were various research groups attempting to identify, characterise and validate Cas9 orthologs derived from bacterial species other than S. pyogenes with the aim of identifying CRISPR/Cas9 systems that were better suited for use in AAV vectors, that were able to target a wider variety of PAM sites, and that also had reduced “off-target” effects (ie. the introduction of unintended breaks).
344 In another paper Wang (2013) received by the publisher in March 2013 and published in Cell in May 2013, the authors state:
There are several potential limitations of the CRISPR/Cas technology. First, the requirement for a NGG PAM sequence of S. pyogenes Cas9 limits the target space in the mouse genome. It has been shown that the Streptococcus thermophilus LMD-9 Cas9 using different PAM sequence can also induce targeted DNA cleavage in mammalian cells (Cong et al., 2013). Therefore, exploiting different Cas9 proteins may enable [sic] to target most of the mouse genome. Second, although the sgRNAs used here showed high targeting efficiency, much work is needed to elucidate the rules for designing sgRNAs with consistent high targeting efficiency, which is essential for multiplexed genome engineering. Third, although our off-target analysis for the seven most likely off targets of Tet1 and Tet2 sgRNAs failed to detect mutations in these loci, it is possible that other mutations were induced following as yet unidentified rules. A more thorough sequencing analysis for a large number of sgRNAs will provide more information about the potential off-target cleavage of the CRISPR/Cas system and lead to a better prediction of potential off-target sites.
345 The paper which was published not long after the priority date was by a research group associated with the Broad Institute. Associate Professor Firestein described the Broad Institute as “a very well-oiled machine … not like a typical academic lab”. He was referring to the breadth of its expertise and the speed with which its research groups could generate data. The paper is consistent with what is in my view the effect of the evidence more generally that the development of new CRISPR/Cas9 systems for use in genome editing using bacterial species that recognised PAM sequences different from those recognised by S. pyogenes and S. thermophilus, would require considerable work in relation to the design of the sgRNA with high targeting efficiency and investigation of the different systems’ off-target effects. I do not consider that the work involved in identifying and characterising systems derived from different bacterial species was likely to have been straightforward or routine. It appears to have been innovative and advanced research work undertaken by highly specialised groups of researchers which included leaders in the field. None of the experts who gave evidence had any experience either individually or as part of a team as at the priority date investigating the use of different CRISPR/Cas9 systems in genome editing or, in particular, the selection or characterisation of bacterial species that might be suitable for that purpose. Of course, that is hardly surprising given the state of the art as at the priority date.
346 On the question of undue burden, ToolGen placed considerable reliance on the Full Court’s decision in Warner-Lambert Co LLC v Apotex Pty Ltd (No. 2) (2018) 129 IPR 205 (“Warner-Lambert FC”), the decision at first instance in the same case in Warner-Lambert, and also the decision of Heerey J in Eli Lilly & Co v Pfizer Overseas Pharmaceuticals (2005) 64 IPR 506 (“Eli Lilly”). Both cases were concerned with methods of treatment using known pharmaceutical compounds, and both were concerned with the application of s 40 of the Act prior to the its amendment by the RTB Act. It was also common ground in both cases that the patent specification in suit contained the information necessary to enable the skilled addressee to prepare some compositions within the claims. In Eli Lilly Heerey J said at [193]:
It would be necessary to test for oral bioavailabilty, toxicity and effectiveness, but the evidence shows that while these steps call for skill, they are essentially routine for those skilled in this area. The term routine here (and in other contexts in this case) is not used as a synonym for simple and easy. In the present case the hypothetical skilled workers at the hypothetical workbench are persons holding academic qualifications at the Ph D [sic] level together with practical experience. It would not be necessary to employ such persons unless the task they had to perform was a difficult one. Yet this does not of itself mean that the patent could not be worked without further invention.
His Honour’s decision on this issue was upheld on appeal: Pfizer Overseas Pharmaceuticals v Eli Lilly & Co (2005) 225 ALR 416 (“Pfizer”) at [342] per French and Lindgren JJ.
347 Pfizer was applied at first instance in Warner-Lambert FC at [262]-[263] which concerned a patent involving a method of treating pain using pregabalin. Dismissing the appeal, the Full Court in Warner-Lambert FC said at [129]:
In this connection, we accept that the appellants raise a valid point of distinction which is relevant to the respondent’s criticism that the specification does not contain, for example, specific dosages or a safety and toxicity profile in respect of the use of the compounds for the treatment of pain in human subjects, and that a clinician would not use the compounds for this purpose without this information. Whilst the primary judge appears to have accepted that, from the clinician’s perspective, this information would be necessary — with the consequence that further work would need to be carried out in this regard before the compound would be put to use in clinical practice — his Honour was not satisfied that this meant that the requirements of s 40(2)(a) of the Act had not been met. This was because, in the circumstances of the present case, the work required to put the invention into practice was of a routine (that is, non-inventive) nature for the person skilled in the art (now accepted on appeal), even though the work to be undertaken would require considerable skill, effort and resources or be complex, time-consuming and expensive.
348 However, it is clear that even if the work required of the skilled addressee is non-inventive and routine, it may still amount to undue burden. The skilled addressee is not expected to engage in an unreasonable amount of experimentation, research or study. If the work required involves “… prolonged study of matters presenting initial difficulty” the claim will not be properly enabled: see Gilead at [438] per Jagot J, upheld on appeal Idenix Pharmaceuticals LLC v Gilead Sciences Pty Ltd (2017) 134 IPR 1 at [144] (considering s 40(2)(a) of the Act before amendment by the RTB Act); see also Mentor Corp. v Hollister Inc [1993] RPC 7 at 13 per Lloyd LJ citing with approval the judgment of Buckley LJ in Valensi v British Radio Corporation [1973] RPC 337 at 377.
349 The routine work referred to in both Eli Lilly and Warner-Lambert FC included work necessary to support an application for regulatory approval. In Merck & Co Inc v Arrow Pharmaceuticals Ltd (2006) 154 FCR 31 at [108] the Full Court observed (although not in the context of sufficiency) that “… it is a matter of notoriety that prolonged testing for the purpose of regulatory approval must occur between the stage of patent application and commercial marketing”. The same point was made by Jacob LJ who, when considering the concept of enabling disclosure, said of genetic engineering and pharmaceutical inventions, “[t]he work that goes into bringing them to market relates to testing efficacy and safety – not in actually making the invented product”: Halliburton Energy Services Inc v Smith International (North Sea) Ltd [2006] EWCA Civ 1715 at [18]. His Lordship also observed that the test of “undue effort” and the words of the relevant statutory provision (“clearly enough and completely enough”) emphasise that the question is one of degree. As to how one is to say when the work involved is too much, his Lordship said at [21]:
The answer is that the line is one to be drawn by an exercise of judgment, taking into account all of the relevant factors, one of which is of course the nature of the invention itself and its field of technology. But there are other factors too – for instance, the width of the patent claim or whether it has functional limitations which require too much work to explore.
350 Besides the width of the claim and any relevant functional limitations, it is also necessary to consider what information has been provided by P1. It is all very well for ToolGen to say that the necessary information forms part of the common general knowledge, but that does not mean that the deployment of the common general knowledge to solve a problem raised in P1 (how to avoid the 5’-NGG-3’ PAM limitation) is routine work that does not amount to an undue burden. P1 provides no direction at all as to what options for approaching and solving the problem exist, how they should be prioritised or whether any of them can be expected to work.
351 The work that the notional skilled team would need to undertake at the priority date to perform the invention of claims 1 and 10 using a bacterial species other than S. pyogenes would in my opinion involve a significant research project that would not be straightforward or routine. My reasons are as follows.
352 First, to the extent that P1 might be understood as inviting the skilled addressee to attempt to perform the invention using Cas9 derived from another species, it offers no relevant encouragement or direction. Nor, as I have previously explained, does P1 disclose any principle of general application.
353 Second, the microbiologist would need to identify one or more suitable candidates. P1 provides no guidance on that topic. If the microbiologist were to rely on the Makarova (2011), the list of potential candidates would number around 120. If the microbiologist used Genebank for this purpose, the list of potential candidates would exceed 1,500. The fact that Professor Giffard chose S. thermophilus (which had been singled out for use in genome editing by Cong et al in early 2013) out of the 1,513 search results generated, seems to have involved a remarkable stroke of luck assuming his selection was not influenced by post priority date developments. Moreover, as previously noted, the S. thermophilus system did not relieve the 3’-NGG-5’ PAM sequence limitation.
354 Third, assuming the microbiologist identified another candidate (eg. Professor Giffard’s S. thermophilus) and determined its endogenous crRNA and tracrRNA sequences, it would be necessary to identify and characterise the mature crRNA and tracrRNA. P1 does not provide any guidance on to how that is to be done. I accept that this task may be within the skill of the microbiologist with expertise in CRISPR/Cas systems, but I do not consider that this would be routine work. There was no evidence to suggest that Professor Giffard had himself performed this task before the priority date.
355 Fourth, the RNA-seq data for most bacterial strains of interest was unlikely to be publicly available at the priority date. At that time, there were only around seven species of bacteria for which Type II repeat spacer expression data had been made available.
356 Fifth, while there were alternative methods of identifying and characterising the relevant sequences (eg. RNA-seq experiment, northern hybridisation/northern blot or RNase protection experiments) that could have been adopted by the skilled team, these depended on obtaining a bacterial isolate for the bacterial species of interest. None of the experts suggested that bacterial isolates for every known species of interest, or even a substantial proportion of them, were publically available at the priority date.
357 Sixth, the use of next generation sequencing needed to perform a RNA-seq experiment and to analyse the cut plasmids generated from the cleavage assays for PAM identification was not something that was routine at the priority date. This was a task that would need to be outsourced to a specialist laboratory.
358 Seventh, the requirement to identify the crRNA and tracrRNA molecules as the first step of a northern hybridisation (northern blot) experiment could be difficult for the reasons explained by Professor Thomas. In particular, it may be difficult to predict with certainty the mature crRNA from the spacer sequences of the CRISPR array because of the processing of pre-crRNA into mature crRNA. It may also be difficult to identify the tracrRNA because it would not be clear what part of the repeat sequence of the CRISPR array would be complementary to the tracrRNA.
359 Eighth, the compilation of a PAM variant library to identify and validate the PAM site was not routine work as at the priority date. None of the molecular biologists had used a PAM variant library at the priority date. Further, if using the in vivo approach proposed by Associate Professor Firestein, cellular repair mechanisms in eukaryotic cells may mutate the cut which would inhibit detection of the PAM. I accept Associate Professor Herold’s evidence that using a barcode approach to overcome this issue would create considerable additional work which would need to be outsourced to a specialist laboratory.
360 Ninth, the alternative method of using an in silico approach for PAM site identification relied on identifying the protospacer of the phage where a particular spacer originated. This method depends on the DNA sequence for the phage being publicly available. Professor Thomas and Associate Professor Herold questioned whether that DNA sequences for all potential phages of interest were publically available at the priority date. Both accepted that, as at the priority date, this would be a matter for the microbiologist to investigate. Professor Giffard did not give evidence on the availability of phage sequences at the priority date. In the absence of evidence to the contrary, it cannot be assumed that the DNA sequence for any particular candidate would have been available. Nor would this method of identifying the PAM site be regarded as routine or straightforward.
361 Tenth, for any Cas9 derived from bacterial species that are not highly homologous to S. pyogenes, the S. pyogenes sgRNA could not be used, in which case Associate Professor Firestein’s third approach of using the endogenous tracrRNA and crRNA to create a sgRNA would need to be adopted. Even if it was possible to use the S. pyogenes sgRNA in another system using Cas9 derived from a different bacterial species, this would not be possible for any Cas9 that was not highly homologous.
362 In my opinion the skilled team would be required to carry out prolonged research and experimentation and would most likely encounter significant difficulties along the way. Much of the work would be non-routine and would be carried out in circumstances where P1 provided no meaningful guidance or direction and no assurance of success.
363 I am persuaded that as at the priority date, P1 did not enable a skilled team including a molecular biologist specialising in genome editing in eukaryotic cells and a microbiologist with expertise in CRISPR/Cas systems in prokaryotes, to make the compositions of claim 1, or perform the methods of claim 10, using a bacterial species other than S. pyogenes, without undue burden.
Engineering S. pyogenes Cas9 to recognise a non-NGG PAM sequence
364 The respondents contended that it would not have been possible for the skilled addressee to engineer Cas9 derived from S. pyogenes, so that it recognised a non-5’-NGG-3’ PAM based on the information in P1 and the common general knowledge at the priority date. Professor Thomas gave evidence, which was not challenged by ToolGen, that the crystal structure of the S. pyogenes Cas9-sgRNA complex with a target DNA and the amino acid residues of S. pyogenes Cas9 involved in PAM recognition were not known in October 2012. He said that, without that structure and the knowledge of what amino acid residues are involved in PAM recognition, it would not be possible for him to engineer those parts of the complex involved in PAM recognition. Associate Professor Herold’s evidence was to a similar effect. He gave evidence, which I accept, that P1 (whether read with or without Jinek) does not provide any guidance as to how to engineer S. pyogenes Cas 9.
365 According to Professor Thomas’ evidence in Thomas 2:
[I]n October 2012 I did not know the crystal structure of the S. pyogenes Cas9-sgRNA complex with a target DNA, and I now know that the crystal structure was not solved and published until 2014. For this reason, I consider that engineering S. pyogenes Cas9 in a manner suggested by P1 in October 2012 would require a considerable amount of complex work by a team of biologists (including a structural biologist). Because the structure of the S. pyogenes Cas9-sgRNA complex with a target DNA was not available in October 2012, the amino acid residues of S. pyogenes Cas9 involved in PAM recognition were not known. Without the structure of the S. pyogenes Cas9-sgRNA complex with a target DNA, it would not be possible to rationally engineer the domain(s) of the Cas9 complex involved in PAM recognition. This would mean that engineering Cas9 by making changes to the S. pyogenes Cas9 amino acid sequence, and then testing the resulting mutants for activity, would involve an immense amount of work, and some luck, including because any changes made could impact the interactions between the Cas9 and the sgRNA (which were also not understood until the structure of the Cas9-sgRNA complex was solved). Alternatively this work would involve first solving the structure of the complex, which requires specialised expertise that is not in the domain of standard molecular biology laboratories.
In light of Professor Thomas’ evidence, which I accept, I find that engineering a S. pyogenes Cas9 to recognise a non-NGG PAM sequence in October 2012 would have involved a significant research effort outside the skill set of the notional skilled addressee. The possibility that there may have been specialist laboratories that may have been engaged to resolve the problem identified by Professor Thomas merely reinforces that conclusion.
Chimeric guide RNA other than sgRNA (+48)
366 Figure 1A in P1 discloses a chimeric guide RNA comprising a crRNA portion fused to a tracrRNA portion which was referred to in the evidence as the “sgRNA (+48)”. This is due to the fact that the sgRNA is truncated at the +48 position of the native S. pyogenes tracrRNA. This truncated form of the tracrRNA, retaining only nucleotides 23 to 48 of the native sequence, is disclosed in Jinek. In JER 1, the experts agreed that P1 does not expressly disclose any chimeric guide RNA other than that depicted in Figure 1A with the truncated tracrRNA.
367 While P1 refers to Jinek, it does so, as I have previously mentioned, by providing background to the genome editing system disclosed in P1 and, in particular, the single-chain chimeric guide RNA. P1 does not provide any information as to how the inventors went about designing the sgRNA (+48) and, in particular, does not provide any guidance or directions enabling the skilled addressee to alter the length of the sgRNA depicted in Figure 1A.
368 In its written submissions, ToolGen relied on oral evidence of Associate Professor Herold and Professor Thomas which was said to support the proposition that the statement in P1 regarding Jinek should not be understood as limiting itself to any particular length of crRNA or tracrRNA that might be used in the single guide as long as the length included the essential portions. The essential portions will comprise the least number of nucleotides of the native tracrRNA sequence necessary for Cas9 mediated cleavage in a eukaryotic cell. P1 does not include any statement to that effect and none can be implied. There is no disclosure in P1 of tracrRNAs of variable length.
369 Further, I did not understand ToolGen to contend that the use of a different sgRNA having a tracrRNA of some different length to that shown in P1 would not involve undue burden if, as I have found, Jinek is neither incorporated by reference, nor common general knowledge. I am satisfied that it would be an undue burden for the skilled addressee (or skilled team if one includes the microbiologist) to redesign the sgRNA without the benefit of the information in Jinek.
370 If I am wrong about my findings in relation to Jinek as not being incorporated into P1 and not being common general knowledge of the skilled addressee (or skilled team if one includes the microbiologist), then I find based on the evidence of Professor Thomas and Associate Professor Herold, which I accept, that developing a longer guide RNA than sgRNA (+48) would involve a significant amount of non-standard work and would involve undue burden even with the assistance of Jinek. The evidence relating to this issue was previously considered in the context of the design and construction of a sgRNA using a bacterial species other than S. pyogenes. Professor Thomas and Associate Professor Herold both agreed that the experimental approach taken by the authors of Jinek, which made use of radiolabeled assays and radioactive gels to test whether or not certain truncated forms of both crRNA and tracrRNA and the Cas 9 cleave double-stranded DNA in vitro, would not have been standard techniques at the priority date and would involve a significant amount of work which could take months. Associate Professor Firestein accepted that he did not have experience with radiolabeled assays at the priority date.
Nuclear localisation sequences (NLSs) and their location
371 A nuclear localisation sequence, or nuclear localisation signal, is a protein (amino-acid sequence) tag that, when added to a protein, “tags” the protein for import into the eukaryotic cell nucleus, across the nuclear membrane, by the cell’s endogenous nuclear transport system. In this case, the NLS will facilitate the entry of the Cas9 protein from the cytoplasm where it is translated into the nucleus of the cell where DNA cleavage occurs.
372 The experts agreed that P1 expressly discloses a particular NLS with the nuclear localisation sequence “PKKKRKV” located at the C-terminus of the Cas9 protein. They also agreed that no other NLS is expressly disclosed. It is common ground that the PKKKRKV NLS was widely used and studied before the priority date, but that there were other NLSs known and used in the art as well. Associate Professor Firestein’s evidence was that he views the sequence and location of the NLS used in P1 as a design choice, and that P1 more broadly discloses the use of any suitable NLS to drive nuclear localisation of the Cas9 protein.
373 ToolGen submitted that P1 implicitly discloses the use of any NLS capable of mediating the entry of the Cas9 protein into the nucleus and is not limited to the PKKKRKV NLS expressly disclosed. The respondents submitted that the choice and position of an NLS can adversely affect the location and function of the protein and is therefore an important matter. They submitted that P1 makes no disclosure of any NLS apart from the PKKKRKV NLS located at the C-terminus.
374 Associate Professor Firestein gave evidence that there are really only 2 positions (the N-terminus and the C-terminus) where the NLS could be located and the testing of those possibilities would be quite straightforward. Associate Professor Herold agreed. Associate Professor Firestein accepted that the position of the NLS could adversely affect localisation and functioning of the protein. In cross-examination he was taken to a post priority date paper concerning the use of Cas9 derived from N. meningitis in genome engineering by Hou et al (2013). He accepted that in the case of N. meningitis Cas9, the paper showed that the protein would not localise in the nucleus with the NLS at either the N-terminus or the C-terminus and that it was necessary for an NLS to be attached to both the C-terminus and the N-terminus if it was to do so. He said that there were other publications that showed that this particular protein can work with a single NLS, but he did not identify the publications or the particular NLS used.
375 I accept Associate Professor Firestein’s characterisation of this aspect of the disclosure. In my view, his interpretation of P1 is analogous to the description of an embodiment of a mechanical device in which a screw is used to fasten two components. To the person skilled in the art this could be fairly understood as a disclosure not only of a screw, but of other forms of suitable fasteners such as a bolt or rivet. I do not think the disclosure of P1 is limited to the PKKKRKV NLS (whether by sequence or location) to what is shown in Figure 1A of P1.
376 I am not persuaded that the skilled addressee would not be able to use another NLS with a different sequence at the same or a different location without undue burden. The evidentiary references provided by the respondents in support of the contrary proposition do not advance its case except perhaps for Associate Professor Herold’s evidence that it could take a month or two to get a different NLS to work. However, Associate Professor Herold and Professor Thomas agreed that this is the type of problem that arises in the work of a molecular biologist and it is dealt with using known techniques. In my opinion, the work associated with the use of a different NLS, positioned at either the C-terminus or N-terminus or at both, would be routine and straightforward and not such as would create undue burden.
Single guide RNA fusion other than with a GAAA linker
377 The crRNA and tracrRNA in the chimeric guide RNA in P1 are fused together with a GAAA linker that forms a hairpin loop as shown in Figure 1A of P1. The experts agreed that P1 does not expressly disclose any linker besides the GAAA linker. However, they also agreed that the GAAA linker is a tool used to connect two RNA molecules and that there is nothing in P1 that excludes the possibility of using another linker. Associate Professor Firestein and Professor Thomas both gave evidence that the nucleotide sequence could be changed but that the length of the nucleotides (i.e. four nucleotides) would need to remain the same. Associate Professor Firestein was of the opinion that P1 more broadly discloses the use of a linker to fuse the crRNA and the tracrRNA sequence in a hairpin loop configuration. I accept Associate Professor Firestein’s characterisation of the relevant disclosure. I think the analogy I previously drew in relation to the disclosure of the NLSis equally applicable here.
378 In my opinion, P1 impliedly discloses to the skilled addressee the use of a suitable linker. The respondents accepted, and I find, that as at the priority date, the skilled addressee would be able to employ a different linker without undue burden.
Use of “paired Cas9 nickases” and Cas9 endonucleases that create staggered-ended double-stranded DNA breaks
379 Given my earlier finding that claims 1 and 10 and their dependent claims do not extend to the use of paired Cas9 nickases, I do not need to consider whether P1 discloses or enables their use. But I should note that ToolGen accepted that P1 does not disclose a system that utilises paired Cas9 nickases.
380 I did find that claims 1 and 10 and their dependent claims include Cas9 endonucleases that create staggered-end double-stranded DNA breaks. The evidence shows that Cas9 derived from Francisella novicida (“FnCas9”), which is used in a CRISPR/Cas9 system described by Chen (2017), creates staggered-end double-stranded DNA breaks. In that system, FnCas9 is complexed with a sgRNA to mediate DNA cutting in eukaryotic cells. Chen (2017) reported that FnCas9 cleaves the target DNA to create four nucleotide long overhands at the 5’ end of each strand. Claims 1 and 10 would cover such a system where it was deployed in vivo using DNA molecules encoding the FnCas9 and the sgRNA.
381 The respondents submitted that P1 does not disclose the use of the FnCas9 system (or any system like it) which produces staggered (or sticky) ended breaks. The three experts accepted that the statement at page 5 of P1 that “RGENs yield blunt ends rather than cohesive ends” meant that the CRISPR/Cas9 system disclosed in P1 produces blunt-ended double-stranded DNA breaks and not staggered-ended double-stranded breaks such as those produced by the FnCas9 based system. I accept that P1 does not disclose a system that produces staggered-ended breaks. The respondents submit that a FnCas9 based system, although within the claims, is not disclosed or enabled by P1.
382 The difficulty I have in relation to the respondents’ submission based on FnCas9 is that the point now taken was not identified in the agreed statement of issues. Although there is discussion in JER 1 concerning Chen (2017), it is directed to a different point (see Question 25). Moreover, the system identified by Chen (2017) is not from the Streptococcus genus and does not provide a basis for finding that a claim based on a Streptococcus Cas9 polypeptide or a S. pyogenes Cas9 polypeptide, would not be enabled or supported by P1. For those reasons I think the respondents should be held to the agreed issues which precludes their reliance on their submissions based on FnCas9 and Chen (2017).
PATENT APPLICATION – DISCLOSURE AND ENABLEMENT
383 The internal disclosure requirement is found in s 40(2)(a) of the Act which states that “a complete specification must disclose the invention in a manner which is clear enough and complete enough for the invention to be performed by a person skilled in the relevant art”. I have previously referred to the Explanatory Memorandum to the RTB Act which states that the new s 40(2)(a) is intended to align the disclosure requirement with that applicable in other jurisdictions which require that the patent application enable the invention to be performed across the scope of the claim: cf. Kimberly-Clark at [25]. I refer to my earlier consideration of the post RTB Act disclosure requirement.
384 There are six points to note in relation to internal disclosure.
385 First, although ToolGen contended that Jinek was incorporated by reference into the patent application, I am satisfied that it is not. As with P1, there is no indication that the authors of the patent application intended that any additional information contained in Jinek beyond what is expressly disclosed should be treated by the reader as incorporated in the patent application whether by reference or otherwise. The information from Jinek referred to in the patent application is no greater (and in fact slightly less) than in P1.
386 Second, the respondents accepted that the patent application, unlike P1, discloses an invention that comprises “a nucleic acid encoding a guide RNA”. They do not contend that the invention of the claims is, in this particular respect, not sufficiently enabled.
387 Third, the patent application differs from P1 in that it discloses the existence of a nucleic acid encoding a Cas9 polypeptide derived from S. pyogenes which in Example 9 is shown to recognise and bind to a 5’-NAG-3’ PAM sequence. It is important to note, however, that Example 9 used Cas9 derived from S. pyogenes. There is no Example in which CRISPR/Cas9 components from other bacterial species are used. The respondents submitted that the patent application does not disclose the invention in each claim in a manner that is clear enough and complete enough for it to be performed by a person skilled in the relevant art because the patent application does not enable an invention comprising a system (or components of a system) derived from a bacterial species other than S. pyogenes without undue burden. I accept that submission essentially for the reasons given in relation to P1. I also accept that the patent application does not enable the use of engineered S. pyogenes derived Cas9 in the compositions or methods of the claims, essentially for the same reasons given in relation to P1.
388 Fourth, ToolGen relied on similar arguments to those which I have previously considered in support of its contention that the length of the sgRNAs disclosed in the patent application are not confined by the single example shown in Figure 1a of the patent application, being a sgRNA (+48). Figure 1a of the patent application reproduces Figure 1A of P1. There is no material difference between the disclosures of P1 and the patent application regarding the design of the sgRNA including its tracrRNA component. In my opinion, the patent application does not provide an enabling disclosure of a sgRNA having a length different from that shown in Figure 1a of the patent application.
389 Fifth, ToolGen relied on similar arguments to those which I have previously considered in support of its submission that the NLSs disclosed in the patent application are not limited to the PKKKRKV NLS and that all other suitable NLSs are implicitly disclosed. I accept that submission. In my opinion, the patent application implicitly discloses an invention that uses any suitable NLS apart from the PKKKRKV NLS both in terms of its sequence and location. I consider that their use would be routine and straightforward, and not such as to create undue burden for the skilled addressee.
390 Sixth, the respondents did not press their ground of opposition based on the disclosure of the GAAA linker in the context of the patent application which makes the same disclosure in Figure 1a of the patent application as is made by Figure 1A in P1.
391 The support requirement is found in s 40(3) of the Act which relevantly provides that the “claims must be supported by the matter disclosed in the specification”. The support requirement in s 40(3) was discussed in the Explanatory Memorandum to the RTB Act. The Explanatory Memorandum states:
Overseas law generally requires there to be a relationship between the claims and the description, and between the claims and any document from which priority is being claimed. This is expressed by the requirement that a claim be ‘supported by’ or ‘fully supported by’ the description. Broadly speaking, the terms ‘support’ and ‘full support’ pick up two concepts:
• there must be a basis in the description for each claim; and
• the scope of the claims must not be broader than is justified by the extent of the description, drawings and contribution to the art.
Despite the underlying concept and policy between fair basis and support being similar, the different terminology has produced different substantive law in different countries.
The difference in substantive law in different countries causes unnecessary complexity and uncertainty for applicants seeking protection in Australia and other jurisdictions. As discussed above (see item 7), having different standards in different countries imposes costs on global innovators, who must familiarise themselves with the varying requirements.
This item is intended to align the Australian requirement with overseas jurisdictions’ requirements (such as the UK). Overseas case law and administrative decisions in respect of the ‘support’ requirement will be available to Australian courts and administrative decision-makers to assist in interpreting the new provision.
392 In Merck Sharp & Dohme Corporation v Wyeth LLC (No 3) (2020) 155 IPR 1 (“Merck”) Burley J, after referring to the relevant extrinsic material (including the Explanatory Memorandum referred to above), observed at [514]:
Having regard to the content of the secondary materials, there can be little doubt that Parliament considers that it is appropriate for the Court to have regard to the law in the European Union and the United Kingdom in considering the scope of the requirement for “support”: Acts Interpretation Act 1901 (Cth) s 15AB.
393 His Honour went on to consider the law in Europe and the United Kingdom concerning Art 84 of the EPC and s 14(5) of the UK Act. Article 84 of the EPC, which is enshrined in s 14(5), provides:
The claims shall define the matter for which protection is sought. They shall be clear and concise and be supported by the description.
Subparagraph (c) of s 14(5) of the UK Act requires that the claims “be supported by the description”.
394 The effect of Burley J’s analysis of the European and UK law is that the “support” requirement or what he called the “claim support obligation” requires that the technical contribution to the art disclosed by the specification justify the breadth of the claim. His Honour referred at [546] to the judgment of Aldous J in Schering Biotech Corp’s Application [1993] RPC 249 (“Schering Biotech”) where his Lordship said at 252-3:
In my view the correct approach under the 1977 Act is to consider the description and claims in the specification through the eyes of the skilled man in the art. Under section 125(1) the invention is that specified in the claims. Thus to decide whether the claims are supported by the description it is necessary to ascertain what is the invention which is specified in the claims and then compare that with the invention which has been described in the specification. Thereafter the court's task is to decide whether the invention in the claims is supported by the description. I do not believe that the mere mention in the specification of features appearing in the claim will necessarily be a sufficient support. The word "support" means more than that and requires the description to be the base which can fairly entitle the patentee to a monopoly of the width claimed. This approach is I believe consistent with the decision of the European Patent Office's Technical Board of Appeals in the Riogen case T301l87 of 16 February 1989 [Biogen NV v Hoffmann-La Roche & Co. AG (Decision T30l/87), [19901 Official Journal E.P.O. 335.]. They said: “The scope of protection in the claims must be fair having regard to the way in which the invention is described and having regard to the information which the skilled person has been given in the description as to how the invention can be carried out”.
Burley J said at [547]:
That approach encapsulates broadly the claim support obligation under s 40(3). To it may be added the requirement that the technical contribution to the art must be ascertained. Where it is a product, it is that which must be supported in the sense that the technical contribution to the art disclosed by the specification must justify the breath [sic] of the monopoly claimed.
395 I agree with Burley J that the approach by Aldous J in Schering Biotech at 252-3 broadly encapsulates the support obligation under s 40(3) of the Act.
396 The question whether a disclosure in a patent application fairly entitles the patentee to a monopoly of the width claimed calls for an assessment of the patentee’s contribution to the art, which must be weighed against the scope of the patentee’s monopoly as defined by the claims. The monopoly, according to UK and European authorities, must be justified by the technical contribution to the art that arises from the disclosure of the specification.
397 At least two difficulties can arise in applying the test. First, as the UK authorities show, determining the patentee’s technical contribution to the art is often not easy. While it has been suggested that the technical contribution may correspond with the inventive step, there will be situations in which this is not so including where, for example, the invention of the claim involves a very significant inventive step and yet, by reason of some deficiency in the disclosure of the specification, the public is deprived of its side of the patent bargain. Leaving aside what the UK authorities sometimes refer to as “classical insufficiency” (broadly corresponding to the requirements of s 40(2)(a) of the Act), this may arise when the specification discloses how to perform the invention across the scope of the claims (eg. by using a known pharmaceutical compound in a new method of treatment) but not the basis upon which the invention of the claim might reasonably be expected to work (ie. by delivering the relevant therapeutic effect). The practice of claiming inventions that are not shown to have a sufficiently plausible or credible justification or support is sometimes referred to as speculative claiming.
398 An important decision in the UK that was directly concerned with the support requirement enshrined in Art 84 of the EPC and s 14(5)(c) of the UK Act is the UK Supreme Court’s decision in Warner-Lambert LLC v Generics (UK) Ltd t/a Mylan [2018] UKSC 56 (“Warner-Lambert UK”) . The leading judgment was given by Lord Sumption (with whom Lord Reed agreed). Much of the discussion in Warner-Lambert UK focused on “second use” claims to new methods of treatment using known pharmaceutical compounds and related Swiss-style claims. But it also includes more general discussion regarding the support requirement albeit in the context of the somewhat different statutory scheme which Burley J elucidated in Merck at [527].
399 Lord Sumption referred at [17] to the “patent bargain” which he described as the foundation of modern patent law both in the UK and the EPO. In this regard, his Lordship referred to the following statement from the decision in EXXON/Fuel Oils (T-409/91) [1994] OJ EPO 653 at paras 3.3 and 3.4 in which the EPO Technical Board observed that it was:
… the general legal principle that the extent of the patent monopoly, as defined by the claims should correspond to the technical contribution to the article in order for it to be supported, or justified. … This means that the definitions in the claims should essentially correspond to the scope of the invention as disclosed in the description. … Although the requirements of articles 83 and 84 are directed to different parts of the patent application, since article 83 relates to the disclosure of the invention, whilst article 84 deals with the definition of the invention by the claims, the underlying purpose of the requirement of support by the description, insofar as its substantive aspect is concerned, and of the requirement of sufficient disclosure is the same, namely to ensure that the patent monopoly should be justified by the actual technical contribution to the art.
400 His Lordship went on to discuss the problem of speculative claiming and the patentee who attempts to claim a monopoly more extensive than could be justified by his or her contribution to the art. This may arise in different contexts including in cases involving claims to wide classes of chemical compounds or cases involving second use patents where known compounds are the subject of claims for methods of treatment or Swiss-style claims directed to new indications. Warner-Lambert UK, which concerned the UK patent for a method of treating pain using pregabalin, was such a case. Lord Sumption, having referred to the discussion in the Court of Appeal’s judgment regarding speculative claiming, said at [22]-[23]:
[22] The Court of Appeal’s reference to “armchair inventors” suggests that what they meant by speculative claiming was claiming by persons who had done nothing new or inventive at all but had simply sought to patent abstract possibilities. That may well be a particular risk in the case of patents for new uses of known compounds, especially when they are commercially successful in their existing use. In reality, however, speculative claiming of this kind is simply one of a number of ways in which a patentee may attempt to claim a monopoly more extensive than anything which is justified by his contribution to the art. Other ways in which this can happen include claiming a monopoly wider than the disclosure in the patent can support. An over-broad claim will not necessarily be speculative. The inventor may really have invented something corresponding to the full breadth of the claim. Research may subsequently demonstrate this. But the claim will still exceed his contribution to the art if that contribution is not sufficiently disclosed in the patent.
[23] The concept of plausibility originates in the case law of the EPO as a response to over-broad claims, in particular claims to whole classes of chemical compounds supported by a description which fails to show which compounds can be expected to work. The Technical Board of Appeal treats the condition of sufficiency under EPC article 83 as satisfied if it is possible to work the invention across the scope of the claim from the information in the specification, interpreted in the light of common general knowledge at the priority date. It addresses the broader question whether the disclosed contribution to the art is commensurate with the monopoly claimed under EPC article 56, in the context of inventive step. In that context, its case law requires the formulation of a problem which the claims of the patent could be said to solve: see T 939/92 AGREVO/Triazole sulphonamides [1996] EPOR 171. It imports a requirement that the patent should disclose not just what the invention is and how to replicate it, but some reason for expecting that it will work. Plausibility was the standard to which the patentee was expected to demonstrate this.
It can be seen that the concept of plausibility has been developed in the UK authorities as a check on speculative claiming and to ensure that the patentee’s monopoly is no more extensive than the contribution to the art made by the relevant disclosure.
401 Lord Sumption referred in some detail to the distinction drawn in the UK cases between so-called “classical insufficiency” (where the skilled person is unable to perform the invention from the information disclosed in the specification) and so-called Biogen insufficiency (where the claim is said to be too broad, because it exceeds the disclosed contribution to the art). The expression Biogen insufficiency is derived from the decision of the House of Lords in Biogen Inc v Medeva Plc [1997] RPC 1. His Lordship said of Biogen insufficiency at [25]:
… The House of Lords imported into section 14(3) of the Act a concept similar to the former requirement of fair basis in section 32(1)(i) of the Patents Act 1949 (“that any claim of the complete specification is not fairly based on the matter disclosed in the specification”). It held that if the claim extended beyond the technical contribution to the art disclosed in the patent, it failed for insufficiency independently of any objection based on want of an inventive step and notwithstanding that the skilled person could perform the invention across the whole scope of the claim. Lord Hoffmann, delivering the leading speech, said at p 50:
But the fact that the skilled man following the teaching of Biogen 1 would have been able to make HBcAg and HBsAg in bacterial cells, or indeed in any cells, does not conclude the matter. I think that in concentrating upon the question of whether Professor Murray’s invention could, so to speak, deliver the goods across the full width of the patent or priority document, the courts and the EPO allowed their attention to be diverted from what seems to me in this particular case the critical issue. It is not whether the claimed invention could deliver the goods, but whether the claims cover other ways in which they might be delivered: ways which owe nothing to the teaching of the patent or any principle which it disclosed.
He went on to make the same point in the context of the objection of insufficiency. Adopting the statement of principle cited above from EXXON/Fuel oils, he pointed out, at p 54, that the purpose of requiring sufficiency of disclosure could not be limited to enabling the public to work the invention after the patent had expired:
Section 72(1)(c) of the 1977 is not only intended to ensure that the public can work the invention after expiration of the monopoly. It is also intended to give the court in revocation proceedings a jurisdiction which mirrors that of the Patent Office under section 14(3) or the EPO under article 83 of the EPC, namely, to hold a patent invalid on the substantive ground that, as the EPO said in Exxon/Fuel Oils (T 409/91) [1994] OJ EPO 653, para 3.3, the extent of the monopoly claimed exceeds the technical contribution to the art made by the invention as described in the specification.
Lord Hoffmann was not, in these observations, addressing the question of second use patents. But such patents raise a similar problem. If it is enough to disclose how to make a known compound and for what conditions, the patentee has acquired a monopoly without adding anything to the sum of knowledge. He will have satisfied the condition of sufficiency but without satisfying its purpose.
402 Lord Sumption set out at [37] a number of propositions relevant to the concept of plausibility. Some of these were expressed in terms most relevant to claims for methods of treatment, Swiss-style claims and suggested therapeutic effects, but they are also relevant to the concept of support more generally. His Lordship observed that the proposition that a product is efficacious for the treatment of a particular condition is not made plausible by a bare assertion to that effect. He went on to consider what information may render an assertion that a product is efficacious plausible. His Lordship observed:
… the claimed therapeutic effect may well be rendered plausible by a specification showing that something was worth trying for a reason, ie not just because there was an abstract possibility that it would work but because reasonable scientific grounds were disclosed for expecting that it might well work. The disclosure of those grounds marks the difference between a speculation and a contribution to the art. This is in substance what the Technical Board of Appeal has held in the context of article 56, when addressing the sufficiency of disclosure made in support of claims extending beyond the teaching of the patent …
403 ToolGen submitted that while claim 1 is ostensibly to a product, namely a Type II CRISPR/Cas system, it is limited by the requirement that it be suitable to make a double-stranded break in a target DNA sequence in a eukaryotic cell. I have already addressed this argument. I do not accept it. The claim is not limited to those compositions capable of making a double-stranded break. Whether or not the composition would make such a break in use would depend on a variety of factors including whether the sgRNA and Cas9 complex was successful in locating and interacting with the target DNA.
404 ToolGen also submitted that the inventors’ technical contribution to the art is, in substance, use of a bacterial Type II CRISPR/Cas system to achieve a double-stranded cut in DNA in a eukaryotic cell using a Cas9 polypeptide from that Type II system with the other components referred to in the claim. It submitted that no one had previously described the use of a CRISPR/Cas system in eukaryotic cells with that ability, and the disclosure of that system represented a substantial contribution to the art. It further submitted that the technical contribution was one of general application and that the width of claims 1 and 10 could be supported on that basis.
405 The respondents submitted that the technical contribution to the art made by the patent application is at best a disclosure of a particular Type II CRISPR/Cas systems derived from S. pyogenes having the following features and characteristics:
(i) a requirement for a 5’-NGG-3’ or 5’-NAG-3’ PAM sequence;
(ii) a chimeric guide RNA having the crRNA and tracrRNA portions identified in Figure 1a a of the patent application and the examples;
(iii) a GAAA linker fusing the crRNA and tracrRNA components in the guide RNA; and
(iv) a particular NLS (PKKKRKV) located at the C-terminus of the Cas9 polypeptide.
They submitted that the scope of the monopoly claimed far exceeds the extent of the contribution to the art given that no Type II CRISPR/Cas9 system is illustrated in the patent application except for the particular system based on S. pyogenes. The last two of these features and characteristics identified by the respondents (i.e. (iii) and (iv)) can be disregarded in light of my previous findings.
406 It is useful to refer back to the patent application and the sections of the specification entitled “Technical Problem” and “Solution of the Problem”. The technical problem disclosed was that a genome editing method for using RNA guided endonuclease based on the CRISPR/Cas system had not been developed. It is then stated that the inventors made many efforts to develop such a system “… and finally established a programmable RNA-guided endonuclease that cleave DNA in a targeted manner in eukaryotic cells and organisms”.
407 The patent application includes statements in [161] that the Cas protein may be isolated from a Streptococcus species, preferably, S. pyogenes, or a recombinant protein, but is not limited to Cas protein so derived. There are also statements in [158] that any Cas protein may be used provided that it has endonuclease or nickase activity when complexed with a guide RNA. The statements that any such Cas protein can be used can be put aside because claim 1 is limited to a composition that uses a Cas9 polypeptide. As I have mentioned, there are no examples given in the patent application of the invention using components derived from any species other than S. pyogenes.
408 In closing submissions ToolGen relied on the decision in Warner-Lambert UK including, in particular, Lord Sumption’s observation at [36] that the test of plausibility is “relatively undemanding” while at the same time expressing a preference for the dissenting judgment of Lady Black. However, ToolGen also stated that it understood the respondents did not rely on any lack of plausibility on the face of the patent application. Having reviewed the respondents’ submissions on this point closely, I think ToolGen’s understanding is correct. This is because the respondents say (as I have already found in relation to P1) that the patent application does not disclose any principle of general application. Accordingly, they say the issue of plausibility does not arise.
409 The respondents submitted that the support requirement in s 40(3) in combination with the requirement of disclosure in s 40(2)(a), operates to ensure that there is an enabling disclosure, and if the disclosure does not enable the invention to be performed to the full extent of the claim, the claim will lack the support required by s 40(3).
410 The facts in Warner-Lambert UK show how a claim might meet the requirement of s 40(2)(a) by providing an enabling disclosure, but not meet the support requirement of s 40(3). However, it is difficult to see how a claim to an invention for which there was no enabling disclosure could meet the support requirement. In such circumstances, the scope of the monopoly defined by the claim could not be justified by the technical contribution to the art. The two requirements are closely interrelated and not wholly distinct in their fields of operation.
411 In the present case, the broadest claims (ie. claims 1, 2, 10 and 11) fail to meet the requirements of both s 40(2)(a) and s 40(3) of the Act due to the fact that they are not confined to the use of Cas9 derived from S. pyogenes. Further, all of the claims, insofar as they encompass other sgRNAs of different lengths to that shown in Figure 1a of the patent application (i.e. sgRNA (+48)), also fail to meet the requirements of both s 40(2)(a) and s 40(3) of the Act.
412 The hearing of the appeal was conducted on the premise that if the claims were not entitled to priority based on P1, then a deferred date of 20 June 2013 would apply (“the deferred priority date”). Although the evidence included two US provisional patent applications filed on 20 March 2013 (P2) and 20 June 2013 (P3), none of the evidence or submissions of the parties made reference to them. P2 is a provisional application entitled “Genotyping with CRISPR/Cas-derived RNA-guided endonucleases” which appears to be solely concerned with a method of using RNA-guided endonucleases in restriction fragment length polymorphism analysis. It does not disclose the invention of any of the claims in the patent application. P3 is a provisional application entitled “Composition for cleaving a target DNA comprising a guide RNA specific for the target DNA and Cas protein-encoding nucleic acid or Cas protein”. The parties did not make any submissions in relation to P3 presumably because it is common ground that even if there are claims entitled to priority based on P3, it was filed too late in time to save the claims of the patent application in light of various journal articles published between the filing of P1 (23 October 2012) and the filing of P3 (20 June 2013).
413 The following publications are relevant to the issues of novelty and inventive step:
Cong et al, “Multiplex Genome Engineering Using CRISPR/Cas Systems” (2013) Science 339, 819-823 and Supplementary Materials;
Mali et al, “RNA-Guided Human Genome Engineering via Cas9” (2013) Science 339, 823-826 and Supplementary Materials; and
Wang et al, “One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering” (2013) Cell 153, 910-918 and Supplementary Information.
414 ToolGen has admitted that Cong (2013) was published on 15 February 2013 and that the Supplementary Materials for Cong (2013) were published on 13 June 2014. However, there is other evidence which shows, and I find, that the Supplementary Materials for Cong (2013) were published on 3 January 2013. ToolGen has admitted that Mali (2013) and its Supplementary Materials were published on 15 February 2013. ToolGen has also admitted that Wang (2013) was published on 9 May 2013 and that its Supplementary Information was published on 2 May 2013.
415 Section 18(1)(b)(i) of the Act provides that an invention is a patentable invention, for the purposes of a standard patent, if the invention, so far as claimed in any claim, when compared with the prior art base as it existed before the priority date, is novel. Section 7(1) of the Act provides that an invention is taken to be novel when compared with the prior art base unless it is not novel in light of relevant prior art information (as defined in the Act).
416 The test for whether a patent claim lacks novelty by reason of a prior publication (or is “anticipated” by a prior publication) has been described as follows in General Tire at 486:
To anticipate the patentee’s claim the prior publication must contain clear and unmistakable directions to do what the patentee claims to have invented … A signpost, however clear, upon the road to the patentee's invention will not suffice. The prior inventor must be clearly shown to have planted his flag at the precise destination before the patentee.
(Citations omitted.)
417 As previously mentioned, ToolGen accepts that if claims 1-8 and 10-18 are not entitled to priority from P1 then the Delegate’s findings that those claims lacked novelty will stand.
418 With regard to claims 9, 19 and 20, the respondents contend that if, contrary to my findings, those claims include the use of in vitro transcribed sgRNA, then they are anticipated by Wang 2013 which discloses systems and methods within those claims.
419 With regard to claims 9 and 20, Associate Professor Herold and Professor Thomas gave unchallenged evidence that T7 promoters were well-known and widely used to drive RNA transcription in vitro and that these produced an RNA molecule with two guanine (G) nucleotides at the 5’ end.
420 There was unchallenged evidence given by Associate Professor Herold in relation to Wang (2013) and its disclosure including the use of a guide RNA with two additional guanine nucleotides at the 5’ end. He said in Herold 1:
The experiments reported in Table 2 that targeted Tet3 were conducted using sgRNA with two extra guanine molecules at the 5' end. I understand this from the bottom of Table S3, which includes the oligonucleotides used to add the T7 promoter to the sgRNA template for in vitro transcription of the sgRNA (see Experimental Section: “Production of Cas9 mRNA and sgRNA” on page 916 and Table S3).
421 ToolGen did not adduce any evidence in answer or make any submission in relation to this evidence. If, as I found, the patent application is not entitled to priority based on P1, and if, contrary to my findings, claims 9 and 20 include the use of in vitro transcribed guide RNA, then those claims, so construed, would lack novelty based on the publication of Wang 2013.
422 Further, if claim 19 (when read with claim 10) does not lack clarity and includes the use of in vitro transcribed guide RNA then that claim would also lack novelty based on the publication of Wang 2013.
423 An invention is a patentable invention if the invention, when compared with the prior art base, involves an inventive step: see s 18(1)(b)(ii) of the Act. Sections 7(2) and (3) of the Act identify the nature of the enquiry. They provide:
(2) For the purposes of this Act, an invention is to be taken to involve an inventive step when compared with the prior art base unless the invention would have been obvious to a person skilled in the relevant art in the light of the common general knowledge as it existed (whether in or out of the patent area) before the priority date of the relevant claim, whether that knowledge is considered separately or together with the information mentioned in subsection (3).
(3) The information for the purposes of subsection (2) is:
(a) any single piece of prior art information; or
(b) a combination of any 2 or more pieces of prior art information that the skilled person mentioned in subsection (2) could, before the priority date of the relevant claim, be reasonably expected to have combined.
424 A “scintilla of invention” can sustain a valid patent, but there must be “some difficulty overcome, some barrier crossed” or something “beyond the skill of the calling”: Lockwood Security Products Pty Ltd v Doric Products Pty Ltd (No 2) (2007) 235 CLR 173 at [52]; RD Werner & Co Inc v Bailey Aluminium Products Pty Ltd (1989) 25 FCR 565 at 574; Allsop Inc v Bintang Ltd (1989) 15 IPR 686 at 701.
425 ToolGen accepts that if claims 1-8 and 10-18 are not entitled to priority from P1 then the Delegate’s finding that those claims lack an inventive step will stand.
426 With regard to claims 9 and 20, I accept it would have been a simple and routine matter for the skilled addressee to use T7 promoters and, by so doing, produce two additional guanine nucleotides at the 5’ end of the guide RNA. Use of T7 promoters would not require any inventive capacity or imagination.
427 Having regard to the unchallenged evidence on this topic, I accept that if, contrary to my findings, the Court was to construe claims 9, 19 and 20 as including the use of in vitro transcribed guide RNA then the evidence shows that these claims would have been obvious in light of each of Wang (2013), Cong (2013) and Mali (2013) taken together with the common general knowledge as at the deferred priority date.
428 On the construction of claims 9 and 20 which limits them to the use of a nucleic acid (such as plasmid DNA) encoding a guide RNA, then the evidence also establishes that each of those claims is obvious in light of each of Cong (2013) and Mali (2013) taken together with the common general knowledge as at the deferred priority date. Cong (2013) and Mali (2013) each describe experiments in which the guide RNA is encoded by a plasmid DNA.
429 Claim 21 introduces the additional step to the method described in any of claims 10-16 wherein the nucleic acid encoding the Cas9 polypeptide is introduced into the eukaryotic cell before introducing the nucleic acid encoding the guide RNA. ToolGen made some very brief oral submissions in relation to claim 21 in which it drew attention to what was said by Associate Professor Herold in Herold 1 concerning claim 21. ToolGen submitted the evidence filed by the respondents does not reach the level required to demonstrate a lack of inventive step for claim 21.
430 The respondents submitted that to introduce the components of the system described in claim 10 in a stepwise fashion would be a simple and routine matter. In their oral evidence each of Professor Thomas, Associate Professor Firestein and Associate Professor Herold agreed that it would be possible to employ the method of the claims using a stepwise approach with a DNA plasmid that encoded for the guide RNA being introduced as a second step. None of the witnesses was asked whether it would be desirable or routine to adopt that approach nor was any of them asked whether it would have been an obvious thing to do.
431 The patent application does not provide any indication that there is anything added by claim 21 to what is claimed in claims 10-20 which could renders claim 21 inventive if (as is the case) none of those claims involves an inventive step. I find that it would be obvious to the skilled addressee that the method of claim 10 (which was itself obvious in light of each of Wang (2013), Cong (2013) and Mali (2013)) could be performed in a stepwise fashion in the manner described in claim 21. Adoption of that method would not require any inventive capacity or imagination or the exercise of skill beyond that of the calling. I therefore find that claim 21 does not involve an inventive step.
432 For the reasons explained each of the claims would, if granted, be invalid. In its closing submissions, ToolGen indicated that it may wish to amend the patent application in the event that I was to find that any one or more of the claims was invalid. Presumably, this would involve amendments aimed at narrowing the scope of the claims and aligning them with the disclosure of P1.
433 The Court has power to hear and determine an application to amend in this case: see s 105(1A) and s 112A of the Act and Meat and Livestock Australia Limited v Branhaven LLC (2020) 281 FCR 640 at [17]-[20], [91]-[93]. ToolGen submitted that it may be appropriate to remit the matter to the Patents Office so that the Delegate could consider the amendment application. I do not think that would be desirable. Given the complexity of this matter, I think ToolGen should make any application to amend the patent application to this Court so that that application may be determined before any final order is made or any application for leave to appeal any such final order is filed. It is plainly desirable that any application to amend the claims be heard and determined in advance of any appeal so that the Full Court may give consideration not only to the issues addressed in these reasons, but any further issues arising out of the foreshadowed amendment application. In this regard, I note that the respondents have already foreshadowed that they will oppose any application that may be made by ToolGen to amend the claims.
434 The only orders I propose to make at this time are procedural orders relating to any application to amend the patent application. In the event that no application to amend is filed within 28 days, I propose to make orders dismissing the appeal and allowing the cross-appeal together with an order directing the Commissioner to refuse the patent application.
435 There does not appear to be any reason why the appellant should not pay the respondents’ costs of the appeal and cross-appeal. I will hear from the parties in relation to costs when the proceeding is next before the Court.
436 Orders accordingly.
I certify that the preceding four hundred and thirty-six (436) numbered paragraphs are a true copy of the Reasons for Judgment of the Honourable Justice Nicholas. |
Annexure A
Bibliography
Bhaya (2011): Bhaya et al, “CRISPR-Cas Systems in Bacteria and Archaea: Versatile Small RNAs for Adaptive Defense and Regulation” (2011) Annu. Rev. Genet. 45 pp 273-297.
Chatterjee (2018): Chatterjee et al, “Minimal PAM Specificity of a Highly Similar SpCas9 Ortholog” (2018) Science Advances 4(10), eaau0766, pp 1-10 and Supplementary Materials.
Chen (2017): Chen et al, “Targeted Activation of Diverse CRISPR-Cas Systems for Mammalian Genome Editing via Proximal CRISPR Targeting” (2017) Nature Communications 8, 14958 pp 1-12 and Supplementary Information.
Chylinski (2013): Chylinski et al, “The tracrRNA and Cas9 Families of Type II CRISPR-Cas Immunity Systems” (2013) RNA Biology 10(5), pp 726-737 and Supplemental Materials.
Cong (2013): Cong et al, “Multiplex Genome Engineering Using CRISPR/Cas Systems” (2013) Science 339(6121), pp 819-823 and Supplementary Materials.
Deltcheva (2011): Deltcheva et al, “CRISPR RNA maturation by a trans-encoded small RNA and host factor RNase III” (2011) Nature 471, pp 602-607.
Fonfara (2013): Fonfara et al, “Phylogeny of Cas9 Determines Functional Exchangeability of Dual-RNA and Cas9 Among Orthologous Type II CRISPR-Cas Systems” (2013) Nucleic Acids Research 42, pp 2577-2590 and Supplementary Tables and Figures.
Grissa (2007): Grissa et al, “CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats” (2007) Nucleic Acids Res. 35, pp 52-57.
Horvath (2010): Horvath et al, “CRISPR/Cas, the Immune System of Bacteria and Archaea” (2010) Science 327(5962), pp 167-170.
Hou (2013): Hou et al, “Efficient Genome Engineering in Human Pluripotent Stem Cells Using Cas9 From Neisseria meningitidis” (2013) Proceedings of the National Academy of Sciences 110(39), pp 15644-15649 and Supporting Information.
Jinek (2012): Jinek et al, “A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity” (2012) Science 337, pp 816-821 and Supplementary Materials.
Kim (2017): Kim et al, “In vivo Genome Editing with a Small Cas9 Orthologue Derived from Campylobacter jejuni” (2017) Nature Communications 8, 14500 pp 1-11 and Supplementary Information.
Mali (2013): Mali et al, “RNA-Guided Human Genome Engineering via Cas9” (2013) Science 339 (6121), pp 823-826 and Supplementary Materials.
Makarova (2011): Makarova et al, “Evolution and classification of the CRISPR-Cas systems” (2011) Nature Reviews Microbiology 9, pp 467-477.
Ran (2015): Ran et al, “In vivo Genome Editing Using Staphylococcus aureus Cas9” (2015) Nature 520, pp 186-191 and Supplementary Information.
Wang (2013) Wang et al, “One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering” (2013) Cell 153, pp 910-918 and Supplementary Information.




