FEDERAL COURT OF AUSTRALIA
Meat & Livestock Australia Limited v Cargill, Inc (No 2) [2019] FCA 33
ORDERS
DATE OF ORDER: |
THE COURT ORDERS THAT:
1. The respondents within 14 days of the date hereof file and serve proposed minutes of orders and short submissions (limited to 3 pages) to give effect to these reasons including the making of final orders concerning the disposition of the appellants’ appeal and the respondents’ amendment application and on any question of costs.
2. The appellants within 14 days of the service of the respondents’ proposed orders and submissions file and serve proposed minutes of orders and short submissions (limited to 3 pages) on such topics.
3. Liberty to apply.
Note: Entry of orders is dealt with in Rule 39.32 of the Federal Court Rules 2011.
BEACH J:
1 On 9 February 2018, I delivered my principal reasons on the appeal brought by Meat & Livestock Australia Limited and another (collectively, MLA); see Meat & Livestock Australia Limited v Cargill, Inc (2018) 354 ALR 95; 129 IPR 278; [2018] FCA 51. I also ordered that the parties file minutes of orders and submissions to give effect to those reasons, including concerning any steps necessary to deal with any application by Branhaven LLC (Branhaven) and Cargill, Inc (Cargill) to amend the claims of the 253 Application which I anticipated that they would make.
2 To recap, the principal claims of the 253 Application involve method claims for identifying a trait of a bovine subject from a nucleic acid sample of that subject. The field of the invention relates to gene association analyses, specifically to single nucleotide polymorphisms and correlated traits of bovine subjects. The scientific disciplines that are relevant are molecular genetics and quantitative genetics. The 253 Application was filed on 1 June 2010 as a divisional application of the parent Australian patent application filed on 31 December 2003, which has now been withdrawn. The parent application claimed a priority date of 31 December 2002 based upon US application number 60/437,482. Accordingly, the co-applicants of the 253 Application, Branhaven and Cargill assert, unsurprisingly, that the earlier priority date applies. In the present context I do not need to add to what I said in my principal reasons concerning the priority date.
3 In my principal reasons I explained why I rejected MLA’s grounds of appeal concerning non-satisfaction of the “manner of manufacture” requirement, lack of novelty, lack of inventive step, lack of sufficiency, lack of fair basis and most of the dimensions of MLA’s lack of utility challenge. But I upheld some of MLA’s grounds concerning lack of clarity and lack of definition, and a residual aspect dealing with lack of utility. I indicated that several integers of the relevant claim(s) would need to be amended to deal with linkage disequilibrium between relevant single nucleotide polymorphisms and also to address questions of statistical significance. I also indicated that if appropriate amendments were made, then my concerns relating to lack of clarity, lack of definition and the residual aspect of lack of utility would fall away.
4 In the circumstances, I refrained from making final orders on MLA’s appeal in order to enable Branhaven and Cargill to consider their position and, if thought appropriate, to apply to amend the relevant claim(s) to address my concerns.
5 On 15 May 2018, Branhaven applied under s 105(1A) of the Patents Act 1990 (Cth) (the Act) to amend various claims of the 253 Application (the amendment application). It purported to do so to address the issues that I had raised in my principal reasons. Cargill was not a co-applicant to the amendment application. I have set out as a schedule to my present reasons the form of these proposed amendments.
6 The amendment application has been supported by evidence adduced by Branhaven from Dr Karen Bentley, a principal of FPA Patent Attorneys and the patent attorney responsible for the prosecution of the 253 Application, Mr Daniel Smith, the intellectual property manager for Branhaven who has been managing its intellectual property portfolio since April 2011, and Professor Jeremy Taylor, a quantitative geneticist who gave expert evidence before me on MLA’s appeal. MLA has relied upon expert evidence from Professor Peter Visscher, a quantitative geneticist, in opposition to the amendment application; he previously gave evidence on the appeal. My principal reasons elaborate on the backgrounds of Professor Taylor and Professor Visscher.
7 Predictably, MLA has opposed the amendment application. Some of the highlights of MLA’s grounds of opposition to the amendment application concern questions of power and are to the following effect:
(a) First, it says that its appeal is no longer on foot in the relevant sense because a final decision has been made by me dealing with all matters in issue. Accordingly, s 105(1A) cannot confer power and jurisdiction where it does not exist. It says that my power and jurisdiction were exhausted when I decided that the claims of the unamended 253 Application would be invalid if granted. And that it makes no difference that final orders have not yet been made.
(b) Second and relatedly, it says that my observations in my principal reasons regarding appropriate amendments do not answer the question of whether I have the jurisdiction to deal with the amendment application.
(c) Third, it says that the Commissioner of Patents’ practice does not answer the question of whether the Court, exercising federal judicial power rather than administrative power, has jurisdiction to entertain an application to amend in the same circumstances or to exercise analogous power.
(d) Fourth, it says that the proposed amendments are not allowable pursuant to s 102(1) of the Act because the amended claims of the 253 Application would claim matter not in substance disclosed in the specification as filed.
(e) Fifth, it says that the proposed amendments are not allowable pursuant to s 102(2)(b) on the ground of lack of fair basis because, as a result of the proposed amendments, the specification of the 253 Application would not comply with s 40(3). In particular, the proposed amendments would add an r2 value for linkage disequilibrium of ≥ 0.7 or ≥ 0.8 in each of the claims and thereby would result in each of those claims not being fairly based on the matter described in the specification because the specification does not describe that linkage disequilibrium should be determined or measured in any particular way. In particular it is said that there is no disclosure in the specification of any method for determining or measuring linkage disequilibrium per se, an r2 value per se, an r2 value as a method for determining or measuring linkage disequilibrium, an r2 value of greater than or equal to 0.7 as a measure of linkage disequilibrium or an r2 value of greater than or equal to 0.8 as a measure of linkage disequilibrium. Further, MLA points out that the specification describes that the degree of linkage disequilibrium varies considerably throughout the genome and is a function of time, recombination events, mutation rate and population structure. Further, MLA highlights that the specification uses distance as a proxy for linkage disequilibrium (see Example 3). Further, it says that the specification uses 500 kilo base pairs (kb) (described as 500,000 nucleotides) either side of a specified SNP as a proxy for linkage disequilibrium.
(f) Sixth, it says that the proposed amendments are not allowable pursuant to s 102(2)(b) on the ground of lack of clarity because, as a result of the proposed amendments, the specification of the 253 Application would not comply with s 40(3). In particular, the proposed amendments would add the word “significantly” before “associated” in each of claims 1, 6, 7, 8, 12 and former claim 14 (now proposed claim 13) and new claims 15, 20, 21, 22, 26 and 27 which include the same phrase “significantly associated”, and thereby would result in each of those claims and claims dependent on them lacking clarity. It is said that it is not clear how the word “significantly” interacts with the added requirement of a particular degree of statistical significance of association identified in each of those claims. Further, it is said that the proposed amendments would add a value of statistical significance of p ≤ 0.05 or of p ≤ 0.01 to each of the claims and thereby would result in each of those claims lacking clarity. It is said that such p-values do not provide a workable standard by which the skilled addressee could determine the boundaries of the claims. In particular, because the p-value will vary depending upon a number of factors including the nature of the population of cattle tested, the skilled addressee would not know whether applying the claimed method to a particular population would infringe the patent. Further, it is said that the proposed amendments would add an r2 value for linkage disequilibrium of ≥ 0.7 or of ≥ 0.8 to each of the relevant claims and thereby would result in each of those claims lacking clarity because such r2 values do not provide a workable standard by which the skilled addressee could determine the boundaries of the claims. In particular, it is said that because the r2 value will vary depending upon a number of factors including the population of cattle tested, the skilled addressee would not know whether applying the claimed method to a particular population would infringe the patent.
(g) Seventh, it says that the proposed amendments are not allowable pursuant to s 102(2)(b) on the ground of lack of definition because, as a result of the proposed amendments, the specification of the 253 Application would not comply with s 40(2)(b) of the Act because the claims would not clearly define the invention for the same reasons I have just described relating to the lack of clarity objection.
8 Now as I have said, each of the above grounds of opposition can best be characterised as a power question. And broadly speaking they can be subdivided into two sub-categories. The first sub-category, which consists of the first three arguments, relates to the question of whether I lack jurisdiction or power to either entertain the amendment application or exercise any power under s 105(1A) because I have already finally determined MLA’s appeal, which determination is said to be embodied in my principal reasons. The second sub-category, which consists of the fourth to seventh arguments, relates to more traditional grounds of objection, namely, whether the proposed amendments are not allowable under ss 102(1) or (2). I will deal with each of these sub-categories of power arguments separately.
9 Now before proceeding further, I would note that there is one other power type argument that is not now pressed in any meaningful sense. Section 105(1A) required the amendment application to be made by the patent applicant(s). In the present case, Cargill and Branhaven were co-applicants of the 253 Application, but the amendment application as instituted had only been made by Branhaven. But on 12 July 2018, Cargill assigned all of its right, title and interest in the 253 Application to SelecTraits Genomics LLC (SelecTraits), a Delaware limited liability company. Further, on 31 July 2018 a copy of the relevant confirmatory assignment deed together with a request to amend ownership details was lodged with IP Australia. Moreover, on 31 July 2018 SelecTraits executed a power of attorney in favour of Branhaven constituting the latter as its attorney to prosecute the amendment application before me on its behalf. On 1 August 2018 I ordered that SelecTraits be added as a party to the proceeding and gave leave to file and serve an amended amendment application reflecting the fact that both Branhaven and SelecTraits were applying to amend the 253 Application. Accordingly, any perceived deficiency on the applicant identity question has now been resolved. For convenience, I will refer to Branhaven in these reasons as also encompassing SelecTraits and its assignor Cargill, unless I indicate otherwise.
10 In addition to the two sub-categories of power questions, MLA has raised various discretionary grounds against granting the amendment application. On the assumption that I have jurisdiction and power, MLA says that I ought not to exercise my discretion pursuant to s 105(1A) in favour of directing the amendment of the 253 Application according to the proposed amendments for the following reasons. First, it says that Branhaven has sought to obtain and has obtained an unfair advantage from the breadth of the claims of the 253 Application in its current form, and it would be unfair for it to now be permitted to amend the 253 Application to narrow those claims. In particular, Branhaven has negotiated to license persons to exploit in Australia the invention the subject of the claims of the 253 Application in their unamended form. Second, it says that Branhaven has unreasonably delayed in seeking to make the proposed amendments. Third, it says that Branhaven was aware or ought to have been aware that the breadth and lack of clarity of the claims of the 253 Application in their unamended form were of great concern to the public including Australian cattle farmers, but persisted in maintaining those claims for as long as it was possible to do so, thereby making unwarranted assertions to the public regarding the potential scope of its monopoly regarding the selection of cattle for breeding. Again to be clear, references to Branhaven include Branhaven and Cargill, with SelecTraits being subject to any burden or impediment to which its assignor was subject and attributed with its assignor’s knowledge.
11 I would also note at this point that MLA’s discretionary arguments proceed on an assumption that Branhaven has challenged. Branhaven contends that absent any s 102 impediment provided by operation of s 105(4), and providing that the power in s 105(1A) has been enlivened, the power in s 105(1A) is not discretionary. In other words I must allow the amendments. Accordingly, it contends that MLA’s discretionary arguments are misconceived. I will dispose of this point later.
12 Further, MLA has asserted that Branhaven elected to persist with the 253 Application in its current form until after I had made my final decision on the appeal and is now bound by its election. Relatedly it says that Branhaven is estopped from bringing the amendment application in accordance with the principles of Anshun estoppel. It says that the issue of the amendment of the 253 Application was so connected with the subject matter of the appeal relating to the unamended 253 Application that it was unreasonable for Branhaven not to have applied to amend the 253 Application during the previous hearing, before I determined all of the issues in the appeal regarding the 253 Application in its current form.
13 In summary, MLA submits that I should refuse the amendment application and that the only appropriate orders I should make given the delivery of my principal reasons on the appeal are that:
(a) MLA’s appeal be allowed;
(b) the decision of the delegate be set aside; and
(c) the 253 Application not proceed to grant.
14 Now given the novelty of some of the legal issues concerning the first sub-category of power questions (non-s 102) and whether the power under s 105(1A) is discretionary, I invited the Commissioner of Patents to make submissions to me on these questions, which the Commissioner did through counsel, Dr Warwick Rothnie, who was of considerable assistance.
15 I would also note one other preliminary matter. The amendment application has been appropriately advertised. On 18 May 2018 I made the following orders:
1. On or before 21 May 2018, the Second Respondent provide to the Commissioner of Patents an advertisement stating the matters identified in r 34.41(1)(a) to (d) of the Federal Court Rules 2011 (Cth) with respect to the amendments to the patent request now sought by the Second Respondent in its interlocutory application filed 15 May 2018 (the amendment application).
2. The Commissioner of Patents publish the advertisement referred to in order 1 in the 31 May 2018 edition of the Official Journal.
3. Any person intending to oppose the amendment application who is not a party to the proceeding and who has notified the parties and the Commissioner of Patents of that intention by 28 June 2018 in accordance with Rule 34.41(1)(d) (third parties), must, by 5 July 2018, file and serve its particulars of grounds of objection to the amendment application.
16 Those orders were complied with. No opposition to the amendment application from any quarter has been forthcoming other than from MLA.
17 In summary, I have decided to reject all of MLA’s principal arguments save the s 105(1A) discretionary power point, and would grant the amendments sought of the 253 Application.
18 For convenience, I have divided my reasons into the following sections:
(a) The progress of the 253 Application ([20] to [37]);
(b) First sub-category of power questions ([38] to [121]);
(c) Some evidence on p-values and r2 ([122] to [193]);
(d) Second sub-category of power questions (s 102) ([194] to [333]);
(e) Discretion ([334] to [466]); and
(f) Conclusion ([467] to [470]).
19 The present reasons are to be read with my principal reasons with defined terms used in my principal reasons equally applying here unless I indicate otherwise. I will also assume a familiarity with the molecular genetics and statistical concepts discussed in my principal reasons and will only elaborate further where necessary. After all, p-values used to reject the null hypothesis and squared coefficients of correlation such as r2 values should be well understood and require only modest elaboration. Generally speaking, the idea, indeed the ideal, is that p-values should be as small as possible to avoid a type 1 error, that is, incorrectly rejecting the null hypothesis or, in other words, to minimise the risk of false positives. The squared coefficient of correlation (r2) should be as high as possible. Let me also be clear at this point that I am not here referring to another capitalised metric R2, which is well known as the coefficient of determination and is used in multiple linear regression analysis. In using the lower case r2 I am here referring to the square of a correlation coefficient “r”; the correlation coefficient “r” can be the Pearson correlation coefficient modified by the Fisher transformation.
THE PROGRESS OF THE 253 APPLICATION
20 It is convenient to begin with a short chronology of the progress of the 253 Application.
21 The 253 Application was filed at the Australian Patent Office on 1 June 2010 as a divisional application of AU2003303599. Examination of the application was requested on 7 December 2010. The first examination report issued on 18 January 2012. No objection to the clarity of the claims, the failure to define the invention or inutility was raised. A second examination report issued on 21 March 2012. No objection to the clarity of the claims, the failure to define the invention or inutility was raised. A third examination report issued on 16 October 2013. No objection to the clarity of the claims, the failure to define the invention or inutility was raised. A notice of acceptance issued on 22 October 2013.
22 A notice of opposition was filed by IP Organisers on 31 January 2014 opposing the grant of the 253 Application. By way of an amendment of notice of opposition dated 17 April 2014, the right on which IP Organisers relied to file the notice of opposition was transferred to MLA.
23 On 30 April 2014, MLA filed a statement of grounds and particulars (the SOGAP). The SOGAP particularised MLA’s allegation of a lack of clarity. An amended SOGAP was filed on 19 December 2015, although no changes were made to the particulars of the allegation of a lack of clarity.
24 Following a hearing held on 1 July 2015, the delegate of the Commissioner of Patents handed down her decision on 6 May 2016 (see Meat & Livestock Australia Ltd and Anor v Cargill, Inc & Anor [2016] APO 26). The delegate concluded that the opposition was only successful concerning a minor issue of clarity relating to claim 13 which Branhaven could overcome by amendment putting to one side the manner of manufacture question concerning claim 13.
25 On 27 May 2016, MLA filed a notice of appeal in this Court from her decision. An amended notice of appeal was filed on 9 September 2016. A further amended notice of appeal was filed on 25 January 2017. A third amended notice of appeal was filed on 15 May 2017. The original notice included a ground of lack of clarity. The amended notice particularised terms alleged to be unclear:
12(b) The following terms in the claims are unclear:
(i) “a method for identifying a trait of a bovine subject”;
(ii) “the at least three SNPs are associated with the trait”;
(iii) “wherein the at least three SNPs occur in more than one gene”;
(iv) “the SNP is about 500,000 or less nucleotides from position 300 of any one of SEQ ID nos 19473 to 21982”.
(Original emphasis.)
26 No amendments to the ground of lack of clarity were made thereafter. There were of course other grounds including lack of definition and also lack of sufficiency which encompassed concerns relating to limb (b) SNPs.
27 On 9 February 2018, I delivered my principal reasons in the appeal from the delegate’s decision. At [944] and [945] of my reasons, I said:
In terms of lack of clarity and for the reasons that I have already expressed in the construction section, claim 1 (and analogous claims) will require amendment to:
(a) define “associated” in terms of statistical significance at the p value of equal to or less than 0.01 (or such other measure as I decide after hearing from counsel further);
(b) require each of the 3 SNPs to satisfy that level of statistical significance (to be discussed further with counsel); and
(c) require the limb (b) SNP to be in LD with the relevant limb (a) SNP and to the requisite degree (to be discussed further with counsel).
As presently formulated, claim 1 and analogous claims fail for lack of clarity and proper definition, and also give rise to aspects of inutility. But if such amendments are made, these matters may be rectified.
28 Branhaven’s amendment application seeks orders for the amendment of the claims of the 253 Application to address my concerns. As I say, I have set out in a schedule to the present reasons the claims with the proposed amendments. In summary, the proposed amendments purport to address the matters referred to in my principal reasons as well as the deletion of claim 13.
29 More particularly, the claims as proposed to be amended may be put into three groups, being claims 1 to 14, claims 15 to 28, and claims 29 to 34. The amendments reflected in these claim groups are as follows.
Claims 1 to 14
30 Claims 1 to 14 are an amended version of existing claims 1 to 15 in the 253 Application.
31 In claim 1 as proposed to be amended:
(a) the words “significantly” and “with the degree of statistical significance being p≤0.05” are inserted so as to define “associated” in terms of statistical significance at that p-value;
(b) the words “each of” are inserted so as to require each of the 3 SNPs to satisfy that level of statistical significance;
(c) the words “and is in linkage disequilibrium with the SNP at position 300 with an r2 value of ≥0.7” are inserted so as to require the limb (b) SNP to be in linkage disequilibrium with the relevant limb (a) SNP to an appropriate degree; and
(d) for ease of reference, the letters “(a)” and “(b)” have been inserted so as to identify those parts of the claim that were referred to at the trial before me and in my principal reasons as “limb (a)” and “limb (b)”.
32 Claim 1 as sought to be amended is in the following marked up form:
1. A method for identifying a trait of a bovine subject from a nucleic acid sample of the bovine subject, comprising identifying in the nucleic acid sample an occurrence of at least three single nucleotide polymorphisms (SNPs) wherein each of the at least three SNPs are significantly associated with the trait, with the degree of statistical significance being p≤0.05, and wherein the at least three SNPs occur in more than one gene; and wherein
and wherein(a) at least one of the SNPs corresponds to position 300 of any one of SEQ ID NOS: 19473 to 21982, or
(b) the SNP is about 500,000 or less nucleotides from position 300 of any one of SEQ ID NOS: 19473 to 21982 and is in linkage disequilibrium with the SNP at position 300 with an r2 value of ≥0.7.
33 Corresponding amendments are made to the similar language in claims 6, 7, 8, 12 and 13 (formerly 14). No amendment has been made to the wording of dependent claims 2 to 5, 9 to 11 and 14 (formerly 15).
34 Further, what was formerly claim 13 is deleted entirely.
Claims 15 to 28
35 Claims 15 to 28 are a new set of claims. They are identical to claims 1 to 14 as proposed to be amended, save that the p-value specified for the degree of statistical significance of the association is 0.01, rather than 0.05. The p-value of 0.01 was, of course, my personal preference as discussed in my principal reasons.
36 It is convenient to set out Claim 15 as sought to be added:
15. A method for identifying a trait of a bovine subject from a nucleic acid sample of the bovine subject, comprising identifying in the nucleic acid sample an occurrence of at least three single nucleotide polymorphisms (SNPs) wherein each of the at least three SNPs are significantly associated with the trait, with the degree of statistical significance being p≤0.01, and wherein the at least three SNPs occur in more than one gene; and wherein
(a) at least one of the SNPs corresponds to position 300 of any one of SEQ ID NOS: 19473 to 21982, or
(b) the SNP is about 500,000 or less nucleotides from position 300 of any one of SEQ ID NOS: 19473 to 21982 and is in linkage disequilibrium with the SNP at position 300 with an r2 value of ≥0.7.
Claims 29 to 34
37 Claims 29 to 34 are a new set of dependent claims. They specify a higher r2 value of ≥0.8 for the degree of linkage disequilibrium between the limb (b) SNP and the relevant limb (a) SNP.
FIRST SUB-CATEGORY OF POWER QUESTIONS
38 It is convenient to now address MLA’s contentions that I do not have jurisdiction to entertain the amendment application or power to grant the amendment sought. I am at this point only dealing with the non-s 102 questions.
(a) MLA’s arguments
39 MLA contends that I do not have the jurisdiction to determine the amendment application or power to grant it.
40 It points out that s 154(1) of the Act confers jurisdiction on the Federal Court with respect to “matters arising under this Act”; see s 39B(1A)(c) of the Judiciary Act 1903 (Cth) and s 76(ii) of the Constitution. Section 154(2) specifically gives the Court jurisdiction to hear and determine appeals against decisions or directions of the Commissioner of Patents.
41 MLA says that the “matter” or justiciable controversy in dispute before me on the appeal was whether the unamended 253 Application could proceed to grant in its current form. This was the subject matter of MLA’s appeal under s 60 of the Act.
42 Now in my principal reasons I upheld MLA’s appeal on the grounds of lack of clarity, failure to define the invention and some aspects of lack of utility. I therefore decided that these grounds of opposition were made out to the requisite high standard required in an appeal from an opposition and that the claims in their unamended form could not proceed to grant. MLA says that my decision that the 253 Application could not proceed to grant with the claims in their original unamended form was a final decision, which dealt with all of the issues arising in its appeal from the opposition so far as they were capable of final determination.
43 MLA made reference to R v Smith; Ex parte Mole Engineering Pty Ltd (1981) 147 CLR 340 at 348 and 349. MLA submitted that in Mole Engineering it was decided that a “decision” of the Commissioner that the claims could not proceed to grant in their current form but could be cured by amendment was a “final decision” for the purposes of an appeal. Similarly here, so MLA contends, my decision concerning the unamended claims is a final decision because the Court, exercising judicial power rather than administrative power, in essence stands in the shoes of the Commissioner in an appeal to the Court from a decision of the Commissioner.
44 Now MLA accepts that final orders disposing of its appeal have neither been pronounced nor entered. Nevertheless, it says that no issues that were in dispute in the appeal remain to be determined. It says that these have been decided for the reasons that I published. In this sense, so MLA submits, I have become functus officio except to the extent necessary to permit the performance of my judicial function to ensure that orders are made and entered to reflect my reasons.
45 Now MLA says that its submission does not detract from the proposition that until the decision is “formally completed”, the Court may amend or reverse the decision. In Smith v New South Wales Bar Association (1992) 176 CLR 256 at 265, Brennan, Dawson, Toohey and Gaudron JJ said:
It has long been the common law that a court may review, correct or alter its judgment at any time until its order has been perfected. … The power is discretionary and, although it exists up until the entry of judgment, it is one that is exercised having regard to the public interest in maintaining the finality of litigation. Thus, if reasons for judgment have been given, the power is only exercised if there is some matter calling for review. And there may be more or less reluctance to exercise the power depending on whether there is an avenue of appeal.
(Citations omitted.)
46 But MLA says that it is well-established that the jurisdiction of the Court to reopen a judgment and to grant a rehearing is to be exercised in “extremely rare” circumstances and with “great caution”, having regard to the importance of the public interest in the finality of litigation; see Wentworth v Woollahra Municipal Council (1982) 149 CLR 672 at 684 cited with approval in Autodesk Inc v Dyason (No 2) (1993) 176 CLR 300 at 302.
47 In the present case, MLA says that there is no application by Branhaven to reopen any part of the decision on the appeal. Put another way, Branhaven necessarily accepts that on the basis of my principal reasons, the unamended 253 Application that was before me on the appeal cannot proceed to grant.
48 MLA says that the issues on the appeal having been determined, and there being no matter calling for review with respect to those issues, I have exhausted my functions with respect to the subject matter of the appeal, save that it remains for orders giving effect to my decision to be made in the exercise of my judicial function.
49 Further, MLA says that, importantly, Branhaven did not file any application to amend the claims of the 253 Application under s 105(1A) before final reasons were delivered. Accordingly, no matter arising under the Act with regard to an amendment was before me in these proceedings.
50 MLA stresses that I do not have jurisdiction or power after making a final decision on the issues in dispute to invite Branhaven to amend the claims of the 253 Application nor to decide such an application. It is said that it was not a matter in dispute before me.
51 Now MLA had to accept that in my principal reasons I said that I would not “make any final orders until Branhaven has been given the opportunity to consider whether to apply to amend any of the claims to address the concerns” (at [948]). But MLA says that once it is recognised that the decision on the issues in dispute (at [945] and [947]) was final and conclusive, the fact that orders have not yet been made does not change the position. Only orders consistent with that decision can be made, namely, that the 253 Application not proceed to grant and that costs be awarded against Branhaven.
52 Let me now turn to another dimension of MLA’s argument on this aspect.
53 Now I accept that s 105(1A) is the only potential source of jurisdiction for me to hear and determine an amendment application in the context of an appeal from a decision of the Commissioner concerning opposition proceedings. But MLA contends that properly construed, s 105(1A) does not provide me with such a jurisdiction. To the contrary, MLA says that the text of s 105(1A) itself, and the background to its introduction, support the conclusion that it is not a source of jurisdiction for the Court to hear and determine an amendment application after the Court has determined all of the issues in an appeal from opposition proceedings, and found that the claims of the patent application cannot proceed to grant because they would not be valid.
54 Section 105(1A) provides as follows:
Order for amendment during an appeal
(1A) If an appeal is made to the Federal Court against a decision or direction of the Commissioner in relation to a patent application, the Federal Court may, on the application of the applicant for the patent, by order direct the amendment of the patent request or the complete specification in the manner specified in the order.
55 MLA says that s 105(1A) gives the Court jurisdiction and accordingly power to hear and determine an amendment application during an appeal from an opposition. But it says that the grant of the power to direct amendment of a patent application is conditioned on the current existence of an appeal that has been made to the Court i.e. on an appeal remaining pending. It is said that this also flows from the introductory words “If an appeal is made …”.
56 MLA says that s 105(1A) assumes that the amendment application was made before or during the hearing of the appeal (i.e. within the period when the appeal is made but not decided), with the amendment application heard and determined before or with the appeal itself. Accordingly, so it is said, the Court could refuse to permit the patent application to proceed to grant in any form that would be invalid. Alternatively, so it is said, the Court could permit the unamended application to proceed to grant (if no ground of opposition were upheld to any extent) or otherwise may direct the amendment of the patent application to overcome any ground of opposition and permit the amended application to proceed to grant. MLA says that such an approach would ensure that “all matters in controversy between the parties may be completely and finally determined and all multiplicity of proceedings concerning any of those matters avoided”, consistently with s 22 of the Federal Court of Australia Act 1976 (Cth).
57 But MLA says that s 105(1A) does not give the Court power expressly or impliedly to hear and determine an application to amend a patent application, which amendment application is made after the Court has heard and determined all of the issues in the appeal from opposition proceedings. More particularly, s 105(1A) does not contemplate a separate hearing and determination of an amendment application made only after the determination of the appeal. Put another way, MLA says that s 105(1A) only gives jurisdiction to the Court in relation to an amendment application made during (within) the appeal. This may be contrasted with s 154(2), which specifically gives the Court jurisdiction to “hear and determine appeals against decisions or directions of the Commissioner”. MLA says that s 105(1A) is a more narrow and specific grant of power dependent upon the currency of an appeal. Section 105(1A) does not give the Court the power to hear and determine an amendment application independently of the appeal against the decision of the Commissioner under s 60(4) of the Act. Accordingly, MLA says that as I have already heard and decided the appeal, and there being no application to reopen the appeal, s 105(1A) has no work to do. On its argument I simply do not have the jurisdiction to hear and determine Branhaven’s amendment application.
58 Furthermore, MLA says that its construction is consistent with the background to the section. Section 105(1A) was introduced into the Act in 2013 by the Intellectual Property Laws Amendment (Raising the Bar) Act 2012 (Cth) (the 2012 Amending Act) specifically to address the situation where amendments were made or proposed to be made to overcome deficiencies identified by the Commissioner after hearing an opposition.
59 MLA says that it is clear from the extrinsic material, which I will discuss later, that s 105(1A) was introduced to deal with the previous situation where the Commissioner found that the claims of a patent application had deficiencies that could be cured by amendment and an application to amend the claims had been made to the Commissioner. In such a case the Court could only deal with an appeal on the unamended specification and did not have the jurisdiction to deal with the amended specification or a proposed amendment to the specification except in a separate appeal against a decision of the Commissioner on the amended claims.
60 Now MLA had to accept that the power conferred by s 105(1A) is not confined to overcoming the deficiencies identified by the Commissioner in the opposition. In particular, it had to accept that s 105(1A) includes the power to amend a patent application to overcome deficiencies identified by the opponent or indeed the Court itself of its own motion during the appeal. This is consistent with one mischief at least which s 105(1A) was intended to fix. But MLA says that s 105(1A) does not provide or encompass a power to amend the 253 Application to overcome my principal decision that the claims would not be valid if granted. MLA says that there is no suggestion in the extrinsic material that s 105(1A) gives the Court the power to consider and decide upon an amendment application made with the purpose of overcoming the Court’s decision that the claims would not be valid if granted. To the contrary, s 105(1A) refers to an amendment application made in the context of an appeal to the Court against a decision or direction of the Commissioner. It does not refer to an amendment application made with the purpose of overcoming an adverse decision of the Court. It says that s 105(1A) can be contrasted with the use of the broader words “in any relevant proceedings in relation to a patent” in s 105(1).
61 Further, MLA says that under the virtue of the construction of s 105(1A) that it extols, s 105(1A) achieves its purpose. When the amendment application is heard and determined with the appeal, the complexity of the appeal process is reduced. Contrastingly, so MLA submits, reading the provision in a way that permits an amendment application to be heard and determined separately from and after the determination of the appeal has the opposite effect. It increases the complexity of the appeal process. It has an effect that is contrary to the statutory purpose.
62 Further, MLA ambitiously asserted that the construction for which Branhaven contends would have the vice of discouraging patent applicants from making an amendment application before or during an appeal, but would rather encourage them to adopt a “wait and see” strategy.
63 Moreover, MLA pushed the envelope even further by saying that if I permit a patent applicant to propose amendments after a final decision, this could lead to the situation where a patent applicant could make applications to amend in perpetuity. It says that, in principle, each time reasons were delivered rejecting a particular set of amendments, a further set of amendments could be proposed. A patent applicant could therefore keep proposing amendments until they were accepted. MLA says that this could not have been the legislative intention in enacting s 105(1A).
64 Further, MLA says that the construction of s 105(1A) for which it contends may be tested in the following way. MLA prays in aid Lord Hoffman’s speech to the House that the “specification is a unilateral document in words of the patentee’s own choosing” and that the “words will usually have been chosen upon skilled advice” (Kirin-Amgen Inc v Hoeschst Marion Roussel Ltd [2005] 1 All ER 667; [2005] RPC 169; (2004) 64 IPR 444; [2004] UKHL 46 at [34]). MLA says that it is up to the applicant to choose the form of the patent application which it propounds, both before the Commissioner and then in any appeal before the Court. MLA says that where issues of potential invalidity by reference to the grounds of opposition have been identified, the patent applicant may seek only to support the unamended specification. Alternatively, the patent applicant may seek to put forward amendments and support those alone, or only if objections are made out. But MLA says that all of this is the patent applicant’s forensic choice which it makes and is required to make before the appeal is decided.
65 MLA says that if issues of invalidity have been identified in the Commissioner’s decision, then the patent applicant is on notice of them. It then must decide what is to be done about them. Alternatively, if the issues of invalidity have been identified for the first time on appeal, whether by the opponent’s grounds of opposition in the Court, or by the Court during the hearing of the appeal, then again the patent applicant is on notice of them. It must then decide what is to be done about them. MLA says that where a patent applicant is on notice of the issues of invalidity, but then stands by and elects to maintain the application in its unamended form instead of seeking amendments either instead of the unamended claims or as a fall back position if objections are made out during the appeal, it cannot later be heard to complain if the decision goes against it.
66 MLA refers to Raleigh Cycle Co Ltd v Miller H & Co Ltd [1951] AC 278 at 291 to 292 where Lord Morton of Henryton said:
in the absence of exceptional circumstances a patentee should not be allowed to amend invalid claims after he has sought to maintain their validity up to this House…
67 Now I would note at this point that this part of Lord Morton’s speech was directed to the question of discretion rather than absence of jurisdiction. And in any event his context is not a relevant analogue. He was dealing with a granted patent and where validity of the original claims had been persisted with right “up to this House”. I am not dealing with either comparable scenario. Moreover, even if I was in an analogous position, no Australian case supports any “exceptional circumstances” threshold, whether under s 105(1) or s 105(1A).
68 More generally, MLA says that the law confines a party to an election that it has made, requires that a party be bound by the conduct of its case and precludes parties in subsequent proceedings from raising causes of action or issues which they could and should have raised in earlier proceedings. MLA says that there is nothing to suggest that the introduction of s 105(1A) was intended to modify this position at general law. In particular, there is nothing to suggest that s 105(1A) was intended to permit a patent applicant to wait until after the determination of an appeal and then apply to amend the patent application in order to overcome the Court’s decision. MLA says that to the contrary, that would involve a radical departure from the position that usually applies to litigants in judicial proceedings.
69 Further, subject to particular exceptions such as statutory provisions permitting the giving of directions to liquidators or receivers or the giving of advice to trustees, MLA says that I ought not give advice to parties in adversarial litigation, but rather I should quell the dispute between them. It says that there is nothing to suggest that s 105(1A) was intended to modify this position. It says that this conclusion is reinforced by s 60(3B), which was also introduced by the 2012 Amending Act. Section 60(3B) provides that “[t]he Commissioner must not refuse an application under this section unless the Commissioner has, where appropriate, given the applicant a reasonable opportunity to amend the relevant specification for the purpose of removing any ground of opposition and the applicant has failed to do so”. But there is no commensurate provision in s 105(1A). But MLA says that this is because it is not needed. It says that patent applicants are subject to the same principles and obligations that apply to all litigants, and a patent applicant should be presumed to act and have acted in the conduct of an appeal to protect its own interests.
70 Now MLA was prepared to entertain the possibility that there may be cases where an issue as to invalidity emerges only during the course of the Court’s consideration of the appeal after hearing but before the delivery of the decision. But it says that this situation is likely to be rare and is not the present case. But in such a case, it says that it may be expected that the Court would bring that issue to the parties’ attention before the determination of the appeal and invite further argument. And if the issue so identified was capable of being overcome by amendment, the patent applicant could then take the opportunity before the appeal was determined to apply for amendment. But MLA says that such a scenario is consistent with its construction of s 105(1A). Moreover, MLA says that if the Court did not bring the issue to the parties’ attention before the delivery of the decision on the appeal such that, on MLA’s construction, the Court would not have the power to entertain an amendment application, then a patent applicant could apply to reopen the appeal. But in such a case the purpose of reopening the appeal (if permitted) would be to permit argument on the issue of invalidity that had not previously been identified and to permit the patent applicant to apply to amend the patent application in accordance with s 105(1A), that is, while the appeal being then reopened on this hypothesis was still on foot. But that is not the present case.
71 Further, for good measure, MLA also threw in the proposition that on the authorities, amendments of granted patents after judgment on validity has been delivered have not been permitted as a matter of procedural fairness.
(b) Analysis
72 Let me first deal with the contention that I am in essence functus officio, to use MLA’s description, because I have made a final decision or determination on MLA’s appeal.
73 First, it should be obvious that I have not yet disposed of MLA’s appeal. No final orders have been made. My principal reasons and the publication thereof are not a judicial act to be characterised as a final decision or determination. Indeed as my principal reasons make clear (see at [15], [944], [946] and [948]), I have refrained from making final orders until any amendment question has been dealt with one way or the other in order to avoid the very consequence for which MLA now contends. No attractive advocacy on Ms Katrina Howard SC’s part can transform my principal reasons into a judicial final order or determination. The reality of the distinction between a court’s reasons for decision on the one hand and the court’s judgment on the other hand is not in doubt. The distinction explains of course why a disappointed litigant can only appeal a judgment (including an order or decree), but not the court’s reasons per se.
74 Second, my jurisdiction to entertain MLA’s appeal was invoked by MLA’s invocation under s 60(4). But at this stage that jurisdiction is still open as I have not formally and finally disposed of MLA’s appeal.
75 Third, in the context of such an appeal and as I have already set out, s 105(1A) provides:
Order for amendment during an appeal
If an appeal is made to the Federal Court against a decision or direction of the Commissioner in relation to a patent application, the Federal Court may, on the application of the applicant for the patent, by order direct the amendment of the patent request or the complete specification in the manner specified in the order.
76 Given that, strictly speaking, MLA’s appeal is still on foot, Branhaven is entitled to make the amendment application and my jurisdiction to entertain that application has been properly invoked. Further, if the heading to s 105(1A) is to be given any weight, the temporal frame of “during an appeal” has not yet terminated.
77 Before dealing with s 105(1A) in detail, let me say something further concerning s 60(4).
78 The nature and scope of the Court’s powers falls to be determined by construing the statutory grant of power in its context and in its historical and constitutional setting. That context now includes ss 105(1A) and 112A, following the passage of the 2012 Amending Act, Sch 3 “Reducing delays in resolution of patent and trade mark applications”, items 6, 7 and 10.
79 In exercising its powers under s 60(4), the Court albeit in the exercise of judicial power is carrying out the same task as the Commissioner and dealing with the same subject matter. The Court stands in the shoes of the Commissioner and exercises the powers and function of the Commissioner to decide the opposition in the context of the appeal before it.
80 In such a context, it is well accepted that in upholding objections to the grant of a patent, the Commissioner in deciding an opposition under s 60(1) should refrain from refusing the application where the objections may be cured by amendment. This is because a decision on an opposition is a final decision only on the issues in the opposition so far as they are capable of final determination at that stage. In Mole Engineering both Mason J (at 349 and 350) and Wilson J (at 355 and 356) referred with approval to passages from:
(a) Richard Fullagar J’s decision in Broken Hill Proprietary Co. Ltd v American CanCo. [1980] VR 143 at 147 that:
the case which the Commissioner is bound by statute ultimately to decide is the opposition proceeding, but I see no reason why the opposition proceeding should not be decided by several decisions provided that each of them can be said in a real sense to decide ‘the case’ as presently constituted so far as that case is at the time susceptible of present decision.
(b) Lloyd-Jacob J’s observations in L Oertling Ltd’s Application for a Patent [1959] RPC 148 at 148 that:
An interim decision, no less than a final decision, conclusively determines the position of the Comptroller-General in relation to such matters as the decision may specify. It is the means whereby the Comptroller notifies the parties of his decision in relation to so much of the dispute as is susceptible of present determination and his decision also as to the manner in which the remaining matters in dispute may be dealt with.
81 Accordingly, although the Commissioner’s decision on an opposition finally determined the issues between the parties on the matters raised in the opposition to that stage subject to any appeal, it did not finally determine the whole opposition if it was appropriate to afford the patent applicant an opportunity to amend. If the patent applicant did not seek to amend, refusal of the application could follow.
82 Now prior to the 2012 Amending Act, the powers of the Court on an appeal from an opposition were arguably more limited than the powers of the Commissioner. One perceived consequence of New England Biolabs Inc v F Hoffman-La Roche AG (2004) 141 FCR 1 at [22] to [50] was that the Court was limited to considering the specification in the form before the Commissioner when deciding the opposition. Unsatisfactorily, this meant that any application to amend the patent application to address objections upheld in the opposition on appeal had to be remitted to the Commissioner to be dealt with.
83 Unsurprisingly, the legislature considered that such a consequence led to inefficiency and unnecessary duplication. Accordingly, s 105(1A) was introduced into the Act by the 2012 Amending Act.
84 The amendment of the Act to include ss 105(1A) and 112A expanded the powers of the Court to deal with the controversy fully and finally. The explanatory memorandum explained (see explanatory memorandum, Intellectual Property Laws Amendment (Raising the Bar) Bill 2011 (Cth), p 76) in relation to the proposed schedule 3 amendments:
Item 6: Patent opposition – amendments directed by the court
[s 105]
This item amends the Patents Act to provide that a court may consider and decide on amendments to a patent application during an appeal from a decision of the Commissioner.
Currently, during an appeal from a decision of the Commissioner the Court must confine itself to the same subject matter as considered by the Commissioner [New England Biolabs Inc v F Hoffman-La Roche AG (2004) 141 FCR 1]. This means that where an applicant has amended their specification subsequent to the Commissioner’s decision, the Court cannot consider the amended specification, even where the amendments may overcome the grounds on which the decision is being appealed.
This adds complexity to the appeals process and to resolution of opposed patent applications.
The item addresses this problem by giving a court power to consider and decide upon any amendments proposed by the applicant while an appeal is on foot. These amendments would be considered under the existing provisions under which courts may direct amendments [Section 105].
The provision applies only to amendment of patent applications, not to amendment of granted patents.
Existing section 105 applies to amendment of patents. It is expected that in exercising their discretion under new subsection 105(1A), the courts will give account to the different factors that are relevant to applications, in contrast to those applying to patents.
(Emphasis added.)
85 This amendment followed from a consultation process undertaken during 2009. It is worth elaborating on that process as the Commissioner’s counsel helpfully did before me as it elucidates in part the genesis of the proposed changes.
86 In June 2009, IP Australia published a consultation paper (Resolving patent opposition proceedings faster: Towards a stronger and more efficient IP rights system, consultation paper, June 2009). In section 3.12 at [92] to [97] the following was stated:
3.12 Amendments directed by the courts
Decisions of the Commissioner in relation to a patent opposition are appellable to the Federal Court of Australia, as are some other decisions of the Commissioner. The Federal Court is able to affirm, reverse or vary the Commissioner’s decision.
After a successful or partly successful opposition, the Commissioner sometimes gives a patent applicant an opportunity to amend the application. For example, even if the application does not validly claim a patentable invention, it might be possible to craft valid claims on the basis of what has been disclosed in the application.
There are two problems relating to appeals of decisions of the Commissioner, principally related to how amendments are dealt with under such appeals, which are of concern to IP Australia.
The first problem is as follows. When an applicant proposes such amendments, there has been some doubt whether the Federal Court is able to consider the appeal on the basis of the amended specification, or whether it has to consider the form of the specification that was considered by the Commissioner, prior to the amendments. Recent case law indicates that the Court is restricted to the latter. This gives rise to inefficiencies in determining appeals of decisions of the Commissioner, and can protract a final resolution of the matter. It would be more practical and efficient if the Court was able to consider an amended specification – the fact that an applicant has proposed amendments indicates a lack of interest on the applicant’s part in pursuing the specification in its form prior to amendment.
The second problem is as follows. There has been conflicting case law in relation to whether the Court is able to direct that a patent application be amended during an appeal. Giving the Court such a power would streamline the appeals process, and would effectively mean that the court could consider the entire appeal, without referring the matter back to IP Australia to consider amendments. This would be similar to the court’s existing power to direct amendment of a patent, and would permit appeals to be dealt with in a less costly and more expeditious manner.
To address this, IP Australia proposes the following changes:
3.12 Proposed change
• The Federal Court would be given the power to direct amendment of an application for a patent, at the request of the patent applicant, during an appeal of a decision of the Commissioner.
• On an appeal of the Commissioner’s decision, the Federal Court would be able to consider any amendments that may have been proposed or made to a specification or patent request since the Commissioner’s decision.
• The Commissioner would not be able to deal with a request for amendment to an application for a patent while an appeal of the Commissioner’s decision in relation to the application is pending before the Federal Court. Any such amendments would have to proceed before the court under its proposed new power.
87 IP Australia received submissions responding to such suggested changes.
88 In November 2009, IP Australia published a further consultation paper (Towards a Stronger and More Efficient IP Rights System, consultation paper, November 2009). In section 3.12 at [112] to [116], referring back to its June proposal, it said the following:
Proposal 3.12 proposed changes to the way the Federal Court deals with amendments to an application which is the subject of an appeal against a decision of the Commissioner. The proposed changes were that:
• the Court would be given the power to direct amendment
• the Court would be able to consider amendments made to the application following the Commissioner’s decision; and
• the Commissioner would not be able to deal with amendments to an application while an appeal of the Commissioner is under appeal.
These changes were intended to clarify and streamline processes in relation to amendments made during an appeal of a decision of the Commissioner.
Submissions generally agreed with the proposals. However one submission disagreed with the proposal that applicants would be unable to prosecute amendments before the Commissioner while the decision of the Commissioner is on appeal.
IP Australia acknowledges that the inability to prosecute amendments before the Commissioner may cause additional costs for parties. However, amendments prosecuted before the Commissioner may be opposed, leading to concurrent proceedings and unnecessary delays. Accordingly, IP Australia considers it better that amendments to applications on appeal from a decision of the Commissioner be dealt with through the Courts. This is analogous to the existing provisions for amendments to granted patents that are subject to court proceedings. IP Australia considers that this could be achieved by amendment of s 105 to include amendments to applications.
It is intended that all amendments to applications on appeal from a decision of the Commissioner be processed through the Court. To this end, a further change is proposed that sets out that a complete specification relating to an application must not be amended, except under s 105, while relevant proceedings in relation to the application are pending. A similar provision presently exists under s 112 for granted patents.
89 As is apparent, the provision which became s 112A was proposed to reinforce and ensure the objective sought to be achieved by the proposed amendment inserting s 105(1A) into the Act.
90 To that end, the explanatory memorandum explained in relation to s 112A:
Item 10: Patent opposition – decisions on appeal
[s 112A]
This item inserts a new provision, section 112A.
This item is consequential upon item 6 above, which gives a court the discretion to consider and decide on amendments to a patent application during an appeal from a decision of the Commissioner.
The item specifies that only the Court can deal with amendments to an application during an appeal to the Court against a decision of the Commissioner relating to that application. This is similar to an existing prohibition, where there are court proceedings in respect of a granted patent [s 112]. The intention is to avoid having the same issues dealt with by different decision makers.
91 In the present case the Court is exercising judicial power for the first time in the controversy between the parties, rather than executive or administrative power. But there is nothing which causes the nature of the task to change.
92 The fact that the Court has decided the issues in the opposition in the context of the appeal as presently constituted before it does not mean that the Court is functus officio. Consistently with the introduction into the Act of ss 105(1A) and 112A, the Court, standing in the shoes of the Commissioner and undertaking the Commissioner’s task of deciding the opposition, has power to consider an application to amend in an appropriate case.
93 The decision from which the appeal to this Court was brought was, as reflected in Meat & Livestock Australia Limited and Anor v Cargill, Inc. and Anor [2016] APO 26 at 2 that:
The opposition succeeds to the extent that claim 13 lacks clarity.
I allow the applicant two months from the date of this decision to propose amendments.
I award costs according to Schedule 8 against the opponent, Meat & Livestock Australia Limited and Dairy Australia Limited.
94 Accordingly, the decision under appeal did not finally resolve all issues between the parties. Subject to any appeal, it decided the opposition only as far as then constituted and afforded the patent applicant an opportunity to amend.
95 Correspondingly and subject to any further appeal, my principal reasons decided the issues in dispute between the parties in the appeal as then constituted. However, my principal reasons contemplated that Branhaven would be given an opportunity to seek to amend, subject to further ruling on the allowability of any proposed amendments.
96 It is a long-established practice in Australian patent law that if the Commissioner or the Court upholds objections to the grant of a patent, the decision-maker will refrain from refusing the application in circumstances where it appears that the objections may be cured by amendment.
97 Mason J acknowledged this practice in Mole Engineering in the context of analogous provisions of the Patents Act 1952 (Cth) as follows (at 348 and 349):
It is a natural consequence of the procedures under Pt V and Pt VIII that an officer who upholds objections to the grant of an application under Pt V will refrain from refusing the application where it appears that the objections may be cured by amendment. Then it is a practical and sensible course to allow the applicant time within which to lodge a request to amend the specification, as Mr Kildea did in this instance. But his decision was nonetheless a final decision on the original unamended application – there was nothing provisional or tentative about the finding on the grounds of objection. It dealt with all the issues arising on the notice of opposition so far as they were capable of final determination.
(Emphasis added.)
98 Contrary to MLA’s contention, the principle to be distilled from this passage and Mole Engineering generally is not that a final decision on an original unamended patent application closes off any opportunity for the patent applicant to amend. Rather the point is that such a decision finally decides all questions of validity arising on the grounds of opposition so far as they are capable of final determination by reference to the form of the patent application as it then stands. It follows that it is not open to the parties and relevantly the opponent to seek to reargue such questions of validity at a later stage by reference to any amended form of the patent application. But such a decision leaves open the prospect that the deficiencies found to exist in the patent application might be addressed by amendment.
99 Indeed, Moshinsky J said in Merial Inc v Intervet International BV (No 4) (2017) 124 IPR 1 at [10] to [25], particularly at [25(d)]:
Where the Commissioner or the Court upholds objections to the grant of a patent, the decision-maker will “refrain from refusing the application where it appears that the objections may be cured by amendment”, such as by the narrowing of the claims set out in the specification: Mole Engineering.
(Emphasis added.)
100 Now the case before me is not like that which was before Moshinsky J, where his Honour declined to grant the applicant an opportunity to amend for the reasons outlined by him at [35] to [36]. His Honour held inter-alia that the applicant was not entitled to the patent by reason of the fact that it did not derive title from the inventor. This was an issue that went to the heart of the patent application and was not capable of being overcome by an appropriate amendment. His Honour considered that the issue of the applicant’s title to the invention had been dealt with generally and finally at the hearing.
101 But the case before me is rather a case of the kind described by Moshinsky J “where grounds of opposition have succeeded in establishing that the specification as it stands is deficient, but the decision-maker (whether it be the Commissioner of Patents or the Court on appeal) decides that the deficiency can be cured by amendments to the specification” (at [34]). I will return to the nature of the power later, whether discretionary or not, and whether the power should be exercised in the case before me.
102 Let me deal with a number of other points.
103 First, s 105(1A) contains no limitation as to the point in time during an appeal at which an amendment application can be made or determined or the relevant amendment power exercised, providing of course that the appeal remains on foot and has not yet been finally disposed of. In such a context, the question of the timing only has relevance to the exercise of the amendment power, not its existence. For completeness and as I have said, the word “during” in the heading to s 105(1A) encompasses any time prior to final disposition of the appeal.
104 Second, any amendment power under s 105(1A) is clearly not confined to addressing adverse findings made by the Commissioner. On its face, s 105(1A) can extend to permitting amendments to address adverse findings by me. And indeed, if I am in essence standing in the shoes of the Commissioner, there would be little reason in logic to distinguish between the two scenarios. Moreover, it is a non-sequitur to argue that because s 105(1A) refers to “an appeal…against a decision or direction of the Commissioner…” that the amendment power is confined to addressing adverse findings of the Commissioner. These words refer to the nature of the appeal itself rather than to confine the scope of the amendment power.
105 Third, if the second point is good, namely that amendments can be made under s 105(1A) to address adverse findings of the Court, where would one find such adverse findings? Why, of course, the Court’s reasons, as in the present case. This demonstrates in another way how flimsy MLA’s arguments are concerning what it describes as functus officio, estoppel etc.
106 Fourth, MLA’s contentions lead to a bizarre result. MLA says that I have no power to make any amendment. Moreover, it says that I have no power to remit the amendment application to the Commissioner; I will discuss this in a moment. Yet if the amendment application had been made in opposition proceedings before the Commissioner, she would have had power to deal with it. But on appeal I am supposed to be standing in the shoes of the Commissioner. So, on the logic of MLA’s argument, I am standing in the shoes of the Commissioner but cannot do the very thing that she could do if the matter were still before her. And even more bizarrely, not even she can now do it. I cannot remit. And if I make final orders at MLA’s insistence that the 253 Application not proceed to grant, there would be no patent application left on foot to amend. MLA’s contentions all lead to a perverse outcome and a significant lacuna in the legislative scheme. This is also a further reason to reject its contention that I have no power to now deal with the amendment application.
107 Fifth, MLA’s approach arguably puts patent applicants in a difficult position. Where an opponent adopts a “kitchen sink” approach and raises numerous grounds of opposition and amends its notice of appeal several times in the course of doing so, a patent applicant would be forced to take the irreversible step of amending its claims prior to the evidence on which those grounds were based being tested at the hearing and prior to the grounds being considered by the Court on the merits. The alternative course would be to abandon any opportunity to amend. This could, in effect, give the opponent a victory on grounds that might ultimately have had little merit, without them being actually determined by the Court. Such a consequence would discourage patent applicants from relying on legitimate and reasonably arguable defences to grounds of opposition, for fear of being found to be wrong and losing the patent application entirely. Moreover, an amendment is generally speaking irreversible, in that once the claims have been narrowed it is not possible to broaden them again. But perhaps the answer to this is that the patent applicant could make a contingent amendment application during the running of the appeal and as a fall back position.
108 Sixth, and contrary to MLA’s submission, I do not consider that CRI026 v Republic of Nauru (2018) 355 ALR 216 at [60] and [63] assists its position. A question in CRI026 concerned the effect of a “corrigendum” to correct a textual error in a tribunal’s reasons. Their Honours said (albeit obiter) at [60]:
where a discretionary power reposed by statute in a decision maker is, upon proper construction, of such a character that it is not exercisable from time to time but rather is spent upon publishing a decision, the decision maker is prevented from later resiling from the decision because the power to do so is spent and the proposed second decision would be ultra vires. … as a general rule, once an administrative tribunal have reached a final decision in respect of a matter before them in accordance with their enabling statute, the decision cannot be revisited because the tribunal have made an error within jurisdiction. ... in such a case, the principle of functus officio applies on policy grounds favouring the finality of proceedings as opposed to the rules of procedure which apply to formal judgments of courts whose decisions are subject to a full appeal. But it is apparent that those observations were directed to the possibility of a statutory tribunal making substantive changes to a decision as the result of a change of mind, substantive error within jurisdiction or subsequent change of circumstances.
(Citations omitted.)
109 But these principles do not deprive me of power to hear the amendment application. Putting to one side that this is a proceeding in the Court, rather than before the Commissioner as an administrative decision maker, I am not being asked to make substantive changes to the conclusions I have already reached in relation to the construction and clarity of the unamended claims. Rather, I am being asked to consider the allowability of amendments consequent upon my earlier findings. In doing so, I am engaging in a separate exercise of power under s 105(1A), rather than re-exercising any spent power.
110 Moreover, as I have already indicated, Mole Engineering does not deny my power to deal with the amendment application. The point in that case was that the matters decided in the hearing of the opposition as then constituted were finally decided and could not be revisited. But the amendment application in the present case does not seek to reopen matters already decided, but to address them on the foundation that they are correct.
111 Seventh, MLA’s federal matter argument may be rejected. The “matter” is the controversy enlivened by the appeal which has not been finally disposed of, the controversy as to whether amendments are allowable, or both.
112 Eighth, MLA’s construction argument that s 105(1A) ought not to be construed so as to allow a radical departure from “common law principles” such as election or estoppel go nowhere. Such questions are adequately dealt with on the question of the exercise of the power, whether discretionary or not, rather than on the question of the existence of the power.
113 Ninth, MLA says that the words “in any relevant proceedings” in s 105(1) give the Court broad powers to amend a patent, including after a decision by the Court that the patent is not valid. By contrast, the opening words of s 105(1A) restrict the power to hearing and determining an amendment application in an appeal from the Commissioner, not after the appeal has been finally decided. If that were intended to be the case, the Act would also have used the words “in any relevant proceedings” in s 105(1A). Now accepting all this to be so, so what? The appeal has not been finally disposed of.
114 Tenth, MLA’s suggestion that one could contingently apply to amend during the hearing of the appeal or before its commencement as a fall back position also has difficulties. It seems to assume that there are only two realistic possibilities for the form of the claim sets: the unamended claim set or an amended claim set that Branhaven might reasonably anticipate was appropriate as a fall back position. But this is unduly narrow. There may be numerous realistic possibilities that could be chosen for the amended claim set as a fall back position. How is Branhaven to know which to choose in advance? And is it seriously suggested that it should put forward multiple contingent amended claim sets as numerous alternative fall back positions to cover itself, such that I would have to deal with all multiple possible alternatives in the one judgment on the appeal and on the amendment application? After all, the hypothesis here is based on MLA’s foundation that I can only deal with the amendment application and appeal together, or that at least I cannot deal with the amendment application after I have ruled in substance on the original claim set. It seems to me that MLA’s position has an air of unreality to it. The Commissioner is empowered to deal with amendments after giving a ruling on the substance of opposition proceedings. I do not see why s 105(1A) should be construed so as to deny me a similar power particularly as in substance I am standing in the shoes of the Commissioner, providing that I have made no final orders on the appeal. Symmetry of context supports symmetry of power, providing that the text of s 105(1A) in context so permits. Of course I accept that the Commissioner can entertain an amendment application at any time, whereas I am circumscribed by s 105(1A). But as I say, the timeframe under s 105(1A) has not yet been exhausted.
115 In my view, whether a patent applicant ought to bring forward amendments during or prior to the hearing of an appeal, as opposed to bringing forward amendments to address objections upheld by a judge after he has addressed them in his reasons, does not go to the existence of power under s 105(1A) but rather whether it should be exercised.
116 Let me deal with one final topic. I have considered whether I could and should remit the amendment application to the Commissioner. But I have decided that I have no power to do so. And even if I did, I do not consider this to be appropriate. Let me explain my reasons.
117 First, the Commissioner has power to deal with the amendment of a patent application pursuant to s 104. But this is subject to s 112A, which precludes the Commissioner from doing so where a relevant appeal to the Court remains on foot. As I have explained, s 112A was introduced into the Act at the same time as s 105(1A), which gave the Court the power to deal with amendments in the course of such an appeal. The statutory scheme as it presently exists thus encourages and requires that amendments in this context will be dealt with by the Court.
118 Second, as Branhaven points out, there have been cases where the Court has suspended its consideration of an appeal such as the present in order to allow the patent applicant the opportunity to amend by way of an application to the Commissioner under s 104. But such cases were prior to the introduction of ss 105(1A) and 112A. Given the latter provision, that practice is no longer possible while such an appeal remains on foot. And in the present case, strictly the appeal before me remains on foot.
119 Third, I raised the question as to whether the Court could finally determine an appeal, and in doing so remit the matter to the Commissioner in order to allow the patent applicant the opportunity to amend. Now the Court’s powers in an appeal, aside from the express power to amend under s 105(1A), are set out in s 160. These include the power to “(d) affirm, reverse or vary the Commissioner’s decision or direction” and “(e) give any judgment, or make any order, that, in all the circumstances, it thinks fit”. But there is no express power to remit the matter to the Commissioner, although it may be that this is encompassed by the broadly stated power to make any order in s 160(e). But in any event, the statutory scheme in its current form, including ss 105(1A) and 112A, evinces an intention that amendments in this context will be dealt with by me.
120 Further, even if it were possible to devise an appropriate formulation to both finally dispose of the appeal and remit the amendment application, such a course would not promote the objectives of efficiency and reduction in duplication and delay sought to be promoted by the insertion of ss 105(1A) and 112A into the Act. I reached the conclusions reflected in my principal reasons on the issues in the appeal on the basis of a substantial body of written and oral evidence not before the Commissioner. An understanding of the evidence and the conclusions I drew from it are necessary for a full and proper understanding of some of the proposed amendments. It would be unduly burdensome on the parties and unnecessarily duplicative to require them to submit that evidence to the Commissioner and to require the Commissioner to analyse and formulate views on that body of evidence. Further, there is also likely to be a risk that the information before the Commissioner may not be as complete as the information before me or that there may be difficulties in the Commissioner’s evaluation thereof who would lack the advantage I had in hearing and evaluating the evidence in real-time during the dynamics of a trial. There would also be a high potential for yet further appeal to the Court from any decision reached by the Commissioner. In these circumstances, remitting the amendment application (if otherwise possible) would not have been appropriate. It would not have promoted efficiency, reduced duplication or led to any faster ultimate resolution.
121 I reject MLA’s first sub-category of power arguments. Let me now turn to considering the evidence on the amendment application as a precursor to discussing the second sub-category of power questions (s 102) and the question of discretion.
SOME EVIDENCE ON P-VALUES AND R-SQUARED
122 As I have noted elsewhere, claims 1, 6, 7, 8, 12 and former claim 14 (now claim 13) have been amended to specify the following:
(a) First, that the at least 3 SNPs are “significantly” associated with the trait, and that “the degree of statistical significance” is defined by way of a p-value of ≤0.05. This is confirmed to be applicable to “each” of the at least 3 SNPs.
(b) Second, that when “the SNP is about 500,000 or less nucleotides from position 300 of any one of SEQ ID NOS: 19743 to 21982”, the SNP “is in linkage disequilibrium with the SNP at position 300” and the extent of linkage disequilibrium is defined by an r2 measure of ≥0.7.
123 Claims 1 to 14 are duplicated as claims 15 to 28 but with a more stringent p-value of ≤0.01. Original claim 13 has been deleted.
124 Claims 29 to 34 are new, and define the r2 measure as ≥0.8.
(a) P-values
125 A p-value is an estimate of the false positive error rate in making decisions based on a summary statistic calculated from the data one has as it relates to the null hypothesis. The p-value is the level of significance at which the null hypothesis is rejected. A type 1 error is a rejection of the null hypothesis when it is in fact true.
126 In the context of the invention described in the 253 Application, the null hypothesis is that a SNP is not associated with a trait. A p-value of 0.05 therefore means that if the relevant experiment was repeated with independent samples of data, then 5% of the samples would lead you to erroneously reject the null hypothesis and conclude the SNP to be associated when the SNP was not associated, that is, a false positive result. Similarly, a p-value of 0.01 means that if the experiment was repeated with independent samples of data, then 1% of the samples would lead you to erroneously reject the null hypothesis and conclude the SNP to be associated when the SNP was not associated.
127 Professor Taylor gave evidence that another way to communicate the same concept is to describe the percentage of true positive associations which is the confidence in the result. The specification refers to the percentage confidence: “A SNP is associated with a trait when at least one nucleotide occurrence of the SNP occurs more frequently in subjects with a certain characteristic of the trait in a statistically significant manner, for example with greater than 80%, 85%, 90%, 95%, or 99% confidence” (at [0051]). But Professor Visscher disagreed with Professor Taylor that the percentage of true associations is the confidence in the results. The rate of true positives in statistical testing is the proportion of all positive tests that are correctly identified as such. It is not the same as the complement of the false positive rate. He said that the 253 Application does not provide any description or other support for the percentage of ‘true’ associations because associations between SNPs and traits were not replicated or validated in an independent experiment. Now I am inclined to agree that Professor Taylor should not have used the concept of “true associations”. But I do accept that the use of “confidence” in the 253 Application can be taken to refer to the probability that a non-associated SNP will be correctly classified by the statistical test as being non-associated. And as to the fact that the associations between the SNPs and traits were not replicated or validated in an independent experiment, I have discussed this in my principal reasons.
128 I am prepared to accept that where the specification refers to 80%, 85%, 90%, 95% and 99% confidence (at [0051]), this can in context be taken as a reference to the complementary p-value of 0.2, 0.15. 0.1, 0. 05 and 0.01 respectively. So, for example, if one takes a p-value of 0.01, there is only a 1% chance that one would get the result if the null hypothesis was true. So the null hypothesis can be rejected. And then making the assumption that the posited hypothesis is the only available alternative to the null hypothesis, one can therefore have 99% confidence that the alternative hypothesis to the null hypothesis is true. On that foundation, the specification therefore directly supports the proposed amendment to a p-value of either ≤0.05 or ≤0.01 as Professor Taylor said.
129 The specification also makes reference to the fact that “[a] statistical analysis result which shows an association of one or more SNPs or haplotypes with a trait with at least 80%, 85%, 90%, 95%, or 99%... confidence, or alternatively a probability of insignificance less than 0.05, can be used to identify SNPs and haplotypes” (at [0080]). Professor Taylor said that the phrase “probability of insignificance” is not a standard term in the field of statistics. However in the context of [80] as a whole, and when expressed as an alternative to having described at least 95% confidence, Professor Taylor understood it to be yet another way to communicate the rate of false positives. That is, 5% of the results will be erroneous. I do not disagree.
130 Further, according to Professor Taylor, the inclusion of a p-value in the proposed claims informed him that the SNPs referred to would be associated with a given trait with 95% confidence (p-value of ≤0.05) or with 99% confidence (p-value of ≤0.01).
131 Now Professor Visscher also accepted that the level of association between a SNP marker and a trait of interest can be measured for any given population in a number of ways. One method is to test each marker for a statistically significant association with a trait of interest applying an appropriate stringency, for example a p-value. And he accepted that a commonly used p-value in statistical testing is 0.05. This means, as I have already indicated, that assuming there is no association between the marker and variation in the trait of interest, the probability that you would observe a statistically significant association due to chance is 5% per marker. But Professor Visscher said that a more stringent threshold should be used when performing a large number of tests because a number of these tests may show an association as a result of chance, and this needs to be taken into account.
132 But I accept Professor Taylor’s evidence that in the context of the claimed method the fact that an association between a SNP and a trait may be detected as a result of chance when performing a large number of tests does not require a more stringent threshold. The issue of false positives does not present a practical issue in practising the claimed method. A validation study would routinely be done in any case where the population of interest was genetically different to the research population. As Professor Taylor said, the SNPs that were most predictive in the population of interest would be chosen as a result of the validation. The validation study would determine which of the SNPs were truly trait associated or were false positives. Any false positives would not be used. He also noted that genomic selection does not require a strong association between all of the utilised SNPs and the trait for which predictions of genetic merit are to be obtained in order to be useful because of the large number of markers that are used. In a similar sense, if false positives were used along with true positives to produce estimates of genetic merit and the effects of all SNPs were simultaneously estimated, the false positives would, on average, have little effect on the estimated genetic merits. I accept that this is so.
133 Now Professor Visscher said that a p-value will depend on various factors including the estimated effect at the SNP and the sample size. The estimated effect at the SNP depends on its linkage disequilibrium (LD) with a quantitative trait locus (QTL) and the effect size at the QTL. LD between the SNP and QTL will generally depend on the physical distance between them, with the association becoming weaker over distance. Furthermore, when using a hypothesis testing approach to test for statistically significant association, the likelihood of identifying a significant association will also depend on the size of the sample in which the association is tested. For example, a statistically significant association between a particular SNP variant and a trait of interest is more likely to be observed in a study of a large population of thousands of cattle than within a smaller test population of several hundred cattle. Finally, any association is breed dependent and is likely to vary between breeds.
134 Professor Taylor agreed with Professor Visscher that the p-value will depend on the sample size. It was well understood by him and other quantitative geneticists (and indeed any scientist) that the magnitude of a p-value is impacted by sample size when the null hypothesis is not true. With any statistical analysis, the smaller the sample size the less is the power to test null hypotheses and so the larger will be the resulting p-value when the null hypothesis is not true. However, it was well understood by him and other population geneticists that in any population genetic study or method, you need to use a sufficiently large sample size to minimise sampling effects (non-representative samples can occur by chance when sample size is small) and provide sufficient power to reject the null hypothesis when it is not true. In particular, in order to know whether a particular SNP was useful as a diagnostic marker, it was understood by him and other quantitative geneticists that you would need to test the association in a reasonably large sample size. The necessary sample size (N) is governed by the true magnitude of the association between the SNP and the trait, which is of course unknown, but also the frequency of the rarer of the two alleles of the SNP in the population (q). And as he said, N should be sufficiently large that the expected number of homozygotes for the rarer allele (Nq2) is at least 10 for SNPs with large associations.
135 Professor Taylor also agreed with Professor Visscher that there are different genotype patterns for different cattle breeds because the SNP allele frequencies (q and 1-q) may differ between breeds, and allele frequencies at the causative locus may also differ. This was well understood by him and other quantitative geneticists. But as Professor Taylor rightly said, any differences in associations between breeds is not relevant because the proposed claims of the 253 Application require a particular level of association and LD to qualify the use of any SNP as a diagnostic. Accordingly any genotypic differences between populations may result in a limb (a) SNP not being used but potentially another limb (b) SNP being used as a diagnostic in that population.
136 Professor Taylor also noted, in my view correctly, that the 253 Application tested for and identified informative limb (a) SNPs using a large number of different breeds of cattle that are commonly used in agriculture world-wide. As such it cast a wide net for SNP discovery which increased the probability that informative SNPs were identified for use in relevant cattle breed populations.
137 I need say nothing further for the moment about p-values.
(b) Quantifying LD (including via r-squared)
138 To recap from my principal reasons and some of the evidence led before me, polymorphisms (such as SNPs) that have alleles that are co-inherited on the same chromosome with a causative polymorphism, that is, a polymorphism that has a direct effect on phenotype, are also referred to as being in potential linkage disequilibrium (LD) with the causative polymorphism. LD is measured as a correlation.
139 LD between two polymorphisms is influenced by several factors, but can be thought of as a function of the time that the respective polymorphisms arose in a given population and the distance between the polymorphisms on the chromosome.
140 As Professor Taylor said in terms of time, the greater the period of time between the respective polymorphisms arising, the lower the level of LD in the population, on average. For example, if a first mutation (polymorphism) arose several thousand years ago in a given bovine population, and the second polymorphism arose only ten years ago in that population, then the two polymorphisms would generally be in very low LD with each other because they will differ greatly in their frequency. Conversely, if two mutations arose at the same time (or close in time) in a population, then, subject to the distance between the two polymorphisms, there would generally be far greater LD between the two polymorphisms because they will be similar in frequency. Similar allele frequencies are required for strong LD between two loci but does not ensure that this will always be the case. Conversely, two loci that differ greatly in their allele frequencies cannot be in strong LD.
141 In terms of distance, the closer the distance between the two polymorphisms on the chromosome the higher will be the LD, on average. This is because the closer together on a chromosome two polymorphisms are, the lower the likelihood of recombination between the sites, and therefore the greater will be the likelihood that allelic combinations on the chromosomes present within the population will be preserved.
142 As Professor Taylor said, these matters were well understood by those in the field of quantitative genetics well before the priority date. This understanding is consistent with what is said in the specification at [0199]: “The degree of LD varies considerably throughout the genome and is function of time, recombination events, mutation rate and population structure.”
143 According to Professor Taylor, he and others like him that work in the field of quantitative genetics refer to LD in terms of ‘strong’ or ‘high’ LD. And he said that despite the terms ‘strong’ and ‘high’ being relative descriptors, they convey a meaning and information. He understood ‘strong linkage disequilibrium’ to mean, inter-alia, that inherently the two SNPs have very similar allele frequencies in a given population.
144 Now LD may be measured. And there were various techniques available to do so before the priority date. The extent of LD could be calculated using algorithms and statistical methods that were well known to Professor Taylor and others in the field of population genetics at the priority date.
145 Now well before the priority date, LD had been commonly measured in terms of a D or D’ value, although r2 which is the squared correlation of alleles at two loci was also utilised. But due to limitations with the D and D’ statistic, r2 has become the more commonly used statistic. As explained in McKay SD et al., “Whole genome linkage disequilibrium maps in cattle” (2007) 8(74) BMC Genetics 1-12, of which Professor Taylor was an author, and albeit after the priority date (at 6):
It has been suggested that when large differences exist between marker allele frequencies, due to the presence of a rare allele, these two measures of LD are divergent. D’ estimates historical recombination through allelic association whereas r2 measures the squared correlation coefficient between locus allele frequencies and is strongly influenced by the order in which the mutations arose (genealogy) and not necessarily the physical distance between loci. In the context of QTL mapping, r2 is the preferred measure of LD, because it quantifies the amount of information that can be inferred about one (perhaps nonobservable quantitative trait or disease) locus from another, and can therefore be used to estimate the number of loci needed for association studies. For this reason we have used r2 as the primary measure of LD in this study.
(Citations omitted.)
146 But according to Professor Taylor, r2 was also used and preferred by some in the quantitative genetics field before the priority date. In a review on LD in humans in Pritchard JK and Przeworski M, “Linkage Disequilibrium in Humans: Models and Data” (2001) 69 Am J Hum Genet 1-14, the authors referred to their use of “one popular measure of LD between pairs of biallelic markers, commonly denoted by r2”. So at 1 and 2 they state:
Models and Measures of LD
LD refers to the nonindependence of alleles at different sites. For example, suppose that allele A at locus 1 and allele B at locus 2 are at frequencies πA and πB, respectively, in the population. If the two loci are independent, then would we expect to see the AB haplotype at frequency πAπB. If the population frequency of the AB haplotype is either higher or lower than this – implying that particular alleles tend to be observed together – then the two loci are said to be in LD.
A wide variety of statistics have been proposed to measure the amount of LD, and these have different strengths, depending on the context. The measurement of LD is a large and complex topic and will not be reviewed in detail here; but see the work of Devlin and Risch (1995); Jorde (2000) and Hudson (2001). Most of the measures of LD that are in wide use quantify the degree of association between pairs of markers. In part, they differ according to the way in which they depend on the marginal allele frequencies. In the present article, we use one popular measure of LD between pairs of biallelic markers, commonly denoted by r2 (elsewhere, r2 is also denoted by Δ2). We also discuss a multilocus approach, based on an underlying population genetic model, that we feel has some advantages as a summary of the overall amount of LD in a region.
Consider two biallelic loci on the same chromosome, with alleles A and a at the first locus and with alleles B and b at the second locus, where the labeling is arbitrary. The allele frequencies will be written as πA, πa, πB, and πb, and the four haplotype frequencies will be written as πAB, πAb, πaB, and πab. Then,
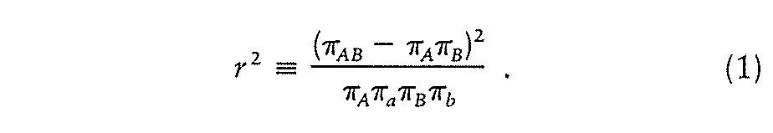
In some of the figures, we plot √r2, because this can make it easier to see the data points. For brevity, we will refer to √r2 as “r”.
In practice, one typically has a sample of m chromosomes from the population. Then, estimates of r2 ( ̂r2) are usually obtained by plugging the sample frequencies ̂πA, ̂πB, ̂πAB, etc., into equation (1).
Besides estimating the amount of disequilibrium between pairs of markers, it is also natural to test the null hypothesis of independence between marker pairs (i.e., linkage equilibrium). This can be done by a x2 test, and it turns out that, for biallelic markers, ̂r2 is the standard x2-test statistic divided by the number of chromosomes in the sample (Weir 1996, p. 113). As shown later in the present article, r2 also arises naturally in the context of association mapping.
The actual value of the disequilibrium coefficient r2 (or any other measure of LD) between two loci is drawn from a probability distribution that results from the evolutionary process. This process can be described in terms of a population genetics tool called the “coalescent” (for reviews, see, e.g., Hudson 1993; Nordborg 2001). When we draw a sample of chromosomes from a population, all the chromosomes are related by some unknown ancestral genealogy, known as a “coalescent tree”. Genetic markers that are very close together on a chromosome have either the same or similar genealogies, and this induces dependence between the alleles at different markers. Markers that are farther apart may have different ancestral genealogies, because of recombination. For this reason, the strength of LD between pairs of markers decreases as a function of the genetic distance between markers.
The expected value of r2 is a function of the parameter p ≡ 4Nec, where c is the recombination rate between the two markers and where Ne is the effective population size. For large p, E(r2) ≈ 1/p (reviewed by Hudson 2001). Below, we will show simulations of the distribution of r under various models.
147 Further, according to Professor Taylor, software programs were also available around 2002 to calculate LD using several different measures including D, D’ and r2. Two programs that he was aware of were Popgene and Merlin. Software programs became more widely available from the mid-2000s.
148 Generally, as the specification described at [0199], LD varies as a function of distance and time. But Professor Taylor said that the degree of LD can be and could be at the priority date readily measured at any time by any of the methods mentioned.
149 Contrastingly, Professor Visscher gave evidence that although he understood r2 values of LD before December 2002, there was no consensus within the field as to the appropriate metric to use to measure LD in cattle. Let me elaborate at this point a little further on r2 before continuing with the discussion concerning LD.
150 A reference to an r value is sometimes referred to as the Pearson correlation coefficient, developed by Karl Pearson. Simply put, it is the covariance of two variables divided by the product of their standard deviations. Related to the use of this coefficient is what was described by some of the experts before me on both the appeal and the amendment application as the Fisher transformation. Because the sampling distribution of the correlation coefficient for highly correlated variables may be significantly skewed, the Fisher transformation converts the skewed distribution of the sample correlation coefficient into a Gaussian distribution. So, the sampling distribution of the correlation coefficient untransformed as I understood it did not conform to the central limit theorem. Solution? Perform the Fisher transformation. Such a transformation thereby enables you to test a hypothesis i.e. to accept or reject the null hypothesis as against the hypothesis of interest, or to provide confidence intervals. Now an r value can range from -1 to +1. It is a measure of the strength of the linear relationship between two variables. A figure of 0 indicates that there is no linear correlation. A figure of 1 indicates a perfect positive linear correlation. A figure of -1 indicates a perfect negative correlation. The value of r may be squared to r2. Values of r2 obviously range between 0 and 1. The closer the value of r2 to 1 the higher the correlation.
151 Contrastingly, R2 is sometimes referred to as the coefficient of determination. The metric is classically used when carrying out a linear regression analysis involving ordinary least squares where one is positing a linear model to explain the relationship between a variable of interest (the dependent variable) and another variable(s) (the independent or explanatory variable(s)). Take a population or sample of interest where you measure the dependent variable and are able to calculate or infer the mean of the variable and the variance from the mean or its standard deviation (the square root of the variance). Now say the dependent variable (y) is plotted on a graph against the explanatory value (x). Say a line of best fit is calculated through the data points using the ordinary least squares method involving a modicum of differential calculus, so that an equation for the line is produced (y = a + bx); I will leave out the error term for the moment. Assuming that the line is a good fit (that concept glosses over a number of issues including avoiding serial correlation, heteroscedasticity etc.), for any measurement of x you can use the equation to predict the y value. But in the real world the observed value of y will usually fall either side of your regression line. So there is a variance of the observed values of y from what the regression line would predict; this is reflected in the residual or error term (c), so that the “true” equation should be y = a + bx + c.
152 Now very simply put, what the R2 metric does is to compare and relate the variance of the dependent variable (y) against its mean on the one hand, with the variance of the dependent variable (y) against the regression line (y being regressed on x). If the variance of the dependent variable against the regression line is much smaller than its variance against the dependent variable’s mean, then the linear model provides a good basis for explaining the variance of the dependent variable from its mean. In simple terms, R2 is calculated as follows:

or (equivalently)

153 The R2 metric is measured between 0 and 1. The higher the R2 value, the greater the variance in the dependent variable from its mean is explained or “determined” by the explanatory variable; hence the expression “coefficient of determination”.
154 Let me make some final points concerning R2 and r2. First, R2 is used in linear regression models as a measure of predictive power. The relationship between specified values of an independent variable(s) (x) and the means of all corresponding values of the dependent variable (y) is modelled for predictive purposes. R2 is the measure of that predictive power. Contrastingly, r2 is used to determine the degree of association or correlation between two variables. It is used to determine the strength of the linear relationship, the present context that I am considering in dealing with LD (I am not at this point discussing predicting phenotype from genotype but the narrower question of the LD between a limb (a) SNP and a limb (b) SNP). Second, it is well accepted that R2 and r2 are not mutually exclusive concepts. Indeed, it can be shown that r2, where you have one independent variable (x) (cf x1, x2… xn) is a special case of R2 making necessary assumptions.
155 Let me return to the question of LD.
156 As should be apparent from what I have just said, LD when measured by r2 is on a continuum from 0 to 1. An r2 value of 0 is indicative of no LD. In other words, the polymorphisms are in linkage equilibrium and therefore knowledge of the identity of an allele at one location tells you nothing more about the identity of an allele at the location of the other polymorphism than was already known based on its frequency in the population.
157 An r2 value of 1.0 is indicative of 100% LD between the polymorphisms. This indicates that knowledge of the allele present at one of the polymorphisms allows you to conclude the presence of the allele at the second polymorphism with 100% accuracy for every chromosome in the population. Consequently, the genotypes of individuals within the given population can be coded in a manner that is identical at both polymorphisms. That is, every animal in the population that is AA, AB or BB, at the first polymorphism will be AA, AB and BB, respectively, at the second polymorphism.
158 Now as I have said, values of r2 are between 0 and 1 and indicative of the degree of LD between the polymorphisms. Professor Taylor understood ‘medium’ or ‘moderate’ LD to equate to an r2 value between approximately 0.3 and 0.7. He understood ‘low’ or ‘weak’ LD to equate to an r2 value greater than 0 but less than 0.3. Contrastingly, although Professor Visscher agreed that the terms “strong” and “high” were relative descriptors of LD, there was no consensus before the priority date (or indeed now) as to any particular value to be ascribed to these relative terms. And he considered Professor Taylor’s attempt to quantify “low”, “moderate” and “high” LD as arbitrary as there were no generally accepted values described as such in the field.
159 Now Professor Taylor said that he and others like him that worked in the field of quantitative genetics well understood at the priority date that there was no need for 100% LD between two polymorphisms such as a SNP and a causal polymorphism, which might itself be a SNP or an insertion or deletion event, in order to detect SNPs that are associated with traits in association analyses. For example, in the trait association research that he has done, the SNPs on the BovineSNP50 assay (which queries 54,001 SNP genotypes in an individual) have an average spacing of about 45,000 bases and have an average r2 value of about 0.2. Moreover, he said that polymorphisms that directly cause trait variation in this context could lie no further than 22,500 bases from a tested marker, and the r2 value between the tested marker and causal variant would, on average, be greater than 0.3. He also explained that the BovineSNP50 assay has been extensively used world-wide for association studies in cattle.
160 Professor Taylor also said that for association studies such as those described in the specification he understood a higher r2 value to be desirable, as the objective of association studies is to generate diagnostic markers that increase the likelihood of correctly identifying the genotype of cattle at polymorphisms that directly cause variation in traits. He said that whilst any degree of LD is better than a random association (i.e. r2 = 0) and is sufficient to improve the accuracy of predictions of cattle genotypes at a polymorphism that directly affects a trait, in practice association studies by their nature and as described in the background section of the specification require higher levels of association to detect causal polymorphisms that might have more modest effect sizes and to increase the degree of predictability of genetic potential in animals for breeding and management decisions.
161 For these reasons, according to Professor Taylor, in the context of association studies, despite the fact that there is no explicit disclosure in the specification of a particular r2 value, he understood the reference to LD in the specification to be, practically, a reference to a need for ‘high’ or ‘strong’ LD, and for that to equate to an r2 value of 0.7 and above. I must say that I have little reason to doubt his evidence or his understanding, which if necessary I am prepared to accept would be the reading and understanding of a person skilled in the art. Further, in [0201] of the specification, there is a reference to LD between the limb (a) and limb (b) SNPs. Professor Taylor’s understanding of the extent of LD that I have just discussed also applies to the extent of LD between the limb (a) and limb (b) SNPs referred to in [0201].
162 Further, Professor Taylor noted that the proposed amendments require that the SNPs in both limbs (i.e. limb (a) and limb (b)) must be significantly associated with a trait. He also noted that the limb (b) SNP retains the requirement to be within 500 kb of the limb (a) SNP.
163 Now Professor Taylor accepted that although the requirement for the respective SNPs to be within 500 kb of each other does not necessarily mean that the two SNPs will be in LD with each other, this distance limitation in the claims, particularly when considered in combination with the requirement that both SNPs must be associated with the same trait, would strongly suggest that the two SNPs were in moderate or strong LD. He pointed out that this is illustrated in Example 3 where the inventors took a sample of the associated SNPs from Example 2 and tested whether additional linked SNPs were associated with the same traits. By analysing the association of SNPs located at varying distances from one of the 2,510 SNPs, the inventors estimated that LD extended about 500 kb. Example 3 provides reasonable (although not conclusive) evidence of LD between the associated SNPs.
164 I must say that I found Professor Taylor’s analysis and opinions on this point and the preceding points persuasive.
165 Now Professor Visscher accepted that if two SNP variants are located close to one another on the genome, the same sets of alleles at each locus tend to travel together from one generation to the next non-randomly and the markers can be considered to be in LD. Where there is essentially complete LD between the two SNPs, trait variation explained by one SNP will be similarly explained by the second SNP. But as the distance between the two SNPs increases, the association of the alleles becomes more random. As with association, the LD measured between two loci also varies between breeds, the population tested and is affected by the population size. For example, the larger the population size the greater the accuracy of the measurement.
166 Professor Visscher accepted that LD can be measured in different ways including the Pearson coefficient of correlation (r), a squared coefficient of correlation (r2), or a standardised relative measure of disequilibrium (D) compared to its maximum expressed as D’.
167 And as he said, LD is not an all or nothing state. In his view it is necessary to specify the strength of the association, which is somewhere between 0 and 1 for both the r2 and D’ values. Using r2 as the measure, zero indicates that two loci are not in LD and therefore a marker at one location tells you nothing about another location. A value of 1 indicates that there is complete LD between two loci and therefore both markers provide the same information in relation to variation in a trait. Further, he said that the precision of estimation will vary with sample size.
168 Now Professor Taylor accepted that the precision of estimation of r2 values, as with any statistical measure, will depend on sample size. Moreover, the hypothesis that r2 is greater or less than certain threshold values such as 0.7 can be statistically tested using the data that were generated to estimate the r2 value. He said that it was understood that a reasonable sample size was required in order for the null hypothesis to be rejected when it is not true.
169 Professor Taylor said that he agreed with Professor Visscher that LD is not an all or nothing state and that the strength of the association ranges from 0 through to 1. This was well understood by him and other population geneticists. But he said that the proposed amended claims specify the particular strength of the required association through the requirement for the LD to be either equal to or greater than an r2 value of 0.7 or, alternatively, 0.8, and these levels are statistically testable in a cattle breed or population.
170 Now Professor Visscher said that the 253 Application did not contain any description or support for a SNP within 500 kb of a specific SNP being in LD with the specific SNP where the extent of LD is an r2 value of equal to or greater than 0.7 or 0.8.
171 Further, he said that the 253 Application did not describe any calculation of LD between markers. There is no discussion or mention in the 253 Application of an r2 value of LD. He said that the 253 Application makes an estimate about the distance over which LD between markers is likely to extend from statistical associations between SNPs and traits based upon the pooled experiments (see Example 3, Determination of the distance of disequilibrium in cattle). But from his reading of the 253 Application, Example 3 is the only relevant discussion of LD.
172 The 253 Application states at [0199] in relation to Example 3 that:
… The degree of LD varies considerably throughout the genome and is a function of time, recombination events, mutation rate and population structure. The extent of LD can vary from a few thousand base pairs to several centimorgans.
173 It states at [0201] that:
… the study was performed to verify the assumption that markers that are in close physical proximity on the bovine genome will associate with the same trait(s) because markers in linkage disequilibrium with the associated SNP marker will also be in linkage disequilibrium with the mutation(s) influencing the trait.
174 As stated in [0208] of the 253 Application, the results of the tests in Example 3 indicate that disequilibrium in cattle exists across the region of 500,000 nucleotides from an associated SNP, in each direction. As such, the 253 Application at [0208] concludes in relation to Example 3 that:
Therefore, it is expected that when an associated SNP is identified, other markers within this 500,000 bp region will also be in disequilibrium with the associated SNP and with the trait of interest, and can be used to infer associations with the trait of interest.
175 But in Professor Visscher’s opinion this is not a measurement of LD, nor does it give any assessment of LD. According to Professor Visscher, there are numerous reasons why two SNPs can be statistically significantly associated with the same trait but not be in LD. Indeed, he said that in order to overcome the fact that a value for LD was not calculated, the 253 Application estimated a distance within which non-specified SNPs are associated with the same trait variant as a specified SNP.
176 It was clear to Professor Visscher from reading Example 3 that the distance 500 kb is used as a proxy for a calculation of LD between markers (based on association with the same trait). However, Example 3 does not provide any information as to the extent of LD, nor what degree of LD is required. According to Professor Visscher, what it does tell is that a SNP within a distance of 500 kb of a specified SNP can be used in place of the specified SNP. But in order for the marker to act as a substitute for a specified SNP and provide the same information in relation to the associated trait as the specified SNP, Professor Visscher expected that the extent of LD, as a measure of r2, would need to be 0.99 or 1. This is because when a SNP is a proxy for another SNP, it will have the same statistical association (p-value) with a trait as the SNP for which it is a substitute. In that sense, they are interchangeable.
177 Now Professor Taylor agreed that the 253 Application did not in terms describe specific methods of calculating LD, including the r2 statistic. However, methods of measuring LD, including the r2 statistic, were well known before the priority date. I accept his evidence as to this.
178 Professor Taylor also disagreed with Professor Visscher that the 253 Application required that the limb (b) SNP must be in complete or virtually complete LD (r2 > 0.99) with the limb (a) SNP. I agree with Professor Taylor.
179 According to Professor Taylor, Professor Visscher’s requirement for an r2 value of 0.99 or 1.0 in effect requires that the limb (b) SNP have genotypes in the population that can be coded to be identical or almost identical to the genotypes of the limb (a) SNP. He says that this misunderstands the purpose of the method described in the 253 Application which is to identify and select desirable traits in cattle through associations between SNPs and the trait. The purpose of the method is not to identify associations between candidate diagnostic SNPs. This is apparent from Example 3 where the inventors took a sample of the associated SNPs from Example 2 and tested whether additional SNPs within 500 kb were associated with the same trait. The purpose of Example 3 was to identify the usefulness of other SNPs within the region (the limb (b) SNPs) to identify and select desirable traits through association. The purpose was not to predict the genotype of the limb (a) SNP. In other words, there is no requirement that the limb (b) SNP be perfectly or nearly perfectly predictive of the limb (a) SNP. However, because the limb (a) SNP is in strong LD with a causal genetic variant that causes trait variation and because the limb (b) SNP is also in strong LD with the same causal variant, Professor Taylor said that one would expect strong, but not perfect or near perfect, LD between the limb (a) SNP and the limb (b) SNP. This is also apparent from the discussion of Example 3 in the 253 Application which indicates that the additional SNPs were identified by reason of their association with the trait and therefore their usefulness in identifying traits of interest, and not by reason of genotypic identity with the limb (a) SNP. I must say that on my own reading of the 253 Application this seems correct.
180 Furthermore, I agree with Professor Taylor that Professor Visscher’s requirement for the limb (b) SNP to be in LD with the limb (a) SNP at an r2 value of 0.99 or 1 fails to recognise that there may be limb (b) SNPs which are not in perfect or near perfect LD with the limb (a) SNP but have a stronger association with the trait than the limb (a) SNP. Such limb (b) SNPs would be useful in identifying trait(s) through association even though they were not genotypically identical or nearly identical to the limb (a) SNP.
181 Now Professor Visscher did agree with Professor Taylor that a high r2 value between a marker and a trait is desirable in association studies and in selection for that trait, but the appropriate level of LD will depend on the context in which the marker is being used. But according to Professor Visscher, a statement of an r2 value of greater than 0.7 or 0.8 was arbitrary as it was not an accepted value in the field and was not described or suggested in the 253 Application.
182 Further, Professor Visscher agreed that the higher the degree of LD between a marker and a causative variant, the more information that will be captured by the marker and its usefulness therefore increases. But according to him, the reference to LD in the proposed amended claims of the 253 Application is between a specified SNP said to be associated with a trait and a non-specified SNP for which no association data is known. He said that this is not the same as the association between a marker and trait discussed in the 253 Application. Professor Visscher understood that the distance of 500 kb was included in the claims as a proxy for LD and in order to capture those SNPs that would be associated with the same trait as a specified SNP. According to his evidence, the non-specified SNP is essentially a substitute (proxy) for a specified SNP and its use is intended to provide the same information in relation to the associated trait as a specified SNP. In that context, in terms of LD, if it is necessary to ascribe an r2 value, he said that he would consider that an r2 value of greater than 0.99 or of 1 would be appropriate.
183 Now Professor Taylor had to agree that an r2 value of greater than 0.7 or 0.8 is, in one sense, arbitrary in the sense that different quantitative geneticists might define slightly different values as implying strong LD. But he said that these r2 values provide a clear threshold which he, and many other quantitative geneticists, would understand as meaning strong LD. That seems to be right in my view.
184 Further, Professor Taylor said that in relation to Professor Visscher’s comment that the requirement for LD is between a trait associated SNP and a SNP for which there is no association data, the proposed amended claims require that the limb (b) SNP be significantly associated with the trait at a p-value of less than or equal to 0.05 or less than or equal to 0.01. The proposed amended claims also require that the limb (a) and limb (b) SNPs have an r2 value of greater than or equal to 0.7 or 0.8.
185 Professor Visscher said that the authors of the 253 Application made no measure of LD. And he therefore did not see how it was possible to select any value for r2 and say that value was based on or supported by what is described in the specification. And as to Professor Taylor’s suggestion that the distance limitation in the claims, when considered in light of the requirement that both SNPs be associated with the same trait, suggests that two such SNPs would be in moderate or strong LD, according to Professor Visscher LD as a function of distance is likely to be ‘low’ over the 500 kb distance required by the claims. He pointed out that in De Roos AP et al, “Linkage Disequilibrium and Persistence of Phase in Holstein-Friesian, Jersey and Angus Cattle” (2008) 179(3) Genetics 1503-1512, on average mean values of r2 were only above 0.8 over distances less than 10 kb, and that at a distance of 500 kb the r2 values were around 0.05 to 0.1. He said that this indicates that there would be only a small degree of LD over distances of 500 kb.
186 But Professor Taylor responded that Professor Visscher’s statement that “LD as a function of distance is likely to be ‘low’ over the 500,000 base pair distance required by [the] claims” reflects what happens ‘on average’ because recombination works to dissipate LD between loci that are separated by large distances on a chromosome. But in Professor Taylor’s view, Professor Visscher’s statement could be simply restated to say that the number of limb (b) SNPs that are in LD with a specific limb (a) SNP at r2 ≥ 0.7 (or r2 ≥ 0.8) will be fewer at a distance of 500 kb than at a distance of 10 kb. But this does not mean that limb (b) SNPs with r2 ≥ 0.7 (or r2 ≥ 0.8) at 500 kb do not exist. It simply means that limb (b) SNPs with r2 ≥ 0.7 (or r2 ≥ 0.8) with the limb (a) SNP will tend to be closer than 500 kb to the limb (a) SNP. I think what Professor Taylor says is correct.
187 Let me move to a slightly different matter. In terms of a lack of clarity, Professor Visscher gave the following evidence.
188 If one requires a marker and a trait to be associated at a specified level of statistical significance, then the level of association detected is dependent on the study design. For example, a marker that is found to be statistically significantly associated with a particular phenotype in cattle for a given population at a p-value of <0.05 may not be significantly associated in a smaller test population or in a different population. The frequency of a particular SNP at particular trait loci will be different between breeds and ultimately the p-value will depend on size and make-up of the test population and other parameters, such as whether there is any adjustment for multiple testing error. And if a population is large enough, it may be possible to detect an association with almost any randomly chosen SNP and a trait. He said that the dependence on factors such as sample size in determining the level of statistical significance means that it is not possible to practically determine whether a particular SNP, or more likely many tens of thousands of SNPs on a SNP chip, has the features of the proposed amended claims without additional information as to how the determination is made. He said that this is particularly so as there is no reference to study design, sample size or a population in the claims and no guidance is provided in the 253 Application.
189 Moreover, he said that a similar issue applies to the calculation of the extent of LD between a non-specified SNP and a specified SNP using a nominated r2 value. The estimate of LD will depend on the sample size and on the particular population tested and the result will also vary by chance. Therefore, if one were to determine that the level of LD between two SNPs was an r2 of 0.7, this is likely only to be true in the particular population measured. But in a population of a different size and different breed makeup, it is likely that one would obtain a different r2 value for the same two SNPs. He said that this difficulty may be overcome by defining the measure in respect of a reference population of a known size and makeup. Now he accepted that while an estimate of LD in a reference population will likewise be dependent on sample size, he said that because the population is known and available to those in the field it can provide a reference point for the assessment of LD. But he said that there is no discussion of a reference population in the claims or in the 253 Application nor was a suitable reference population available before December 2002. He said that the claims do not specify the size of the population in which the association or extent of LD is to be measured nor, for example, whether the assessment is to be carried out in the population where the selection is to be applied or a reference population. He said that the assessment of both of these features is dependent on population size such that different sized populations are likely to yield differing results for association between a SNP and a trait or LD between two SNPs. Further, he said that the association and LD features will be affected by the breed of the population in which the assessment is carried out unless any two SNPs are in LD with an r2 value of 1 (or 0.99). This is because LD at such high r2 values is likely conserved across breeds, that is, identical estimates are likely to be obtained in different breeds of cattle. So it was that he said that for a non-specified SNP to be useful as a substitute for a specified SNP across breeds, an r2 value of between 0.99 and 1 should be specified for the extent of LD.
190 But Professor Taylor responded that there were many association studies (albeit not genome-wide) between genetic markers and traits conducted before the priority date, both in relation to cattle and other species including humans. In all of these studies, the methodology including the sample size was designed so that meaningful and useful associations could be detected.
191 Professor Taylor also responded that the population that is important for the characterisation of the extent of LD is the population that the method is to be used in. In other words, if the method were to be used to identify traits in Herefords, then the relevant population for which LD of r2 ≥ 0.7 (or r2 ≥ 0.8) would be required between limb (b) SNPs and a specific limb (a) SNP would be Herefords. If the method were to be used to identify traits in Holsteins, then the relevant population for which LD of r2 ≥ 0.7 (or r2 ≥ 0.8) would be required between limb (b) SNPs and a specific limb (a) SNP would be Holsteins. This must be right. He also said that the use of an artificial ‘reference population’ could easily lead to misleading results because the reference population would be a mixture of individuals from several breeds and would be unlikely to be representative of the patterns of LD in the actual population in which the method is to be used. I agree. I do not accept that the claims need to specify a reference population as to size or breed.
(c) General
192 Both Professor Taylor and Professor Visscher are highly skilled quantitative geneticists. And to some extent it was difficult to judge between their differing opinions. But it seemed to me that a difference between them was that although both had distinguished academic careers, Professor Taylor also had more practical industry experience in the last 20 years in the field or at least the edge in that respect than Professor Visscher.
193 For my part as a trial judge, Professor Taylor’s views sat more easily with me as being deeply informed by his practical experience in the field in the areas of interest. His views also seemed to square more readily with what I would consider to be a practical reading of the specification by a person skilled in the art. Accordingly, I have preferred Professor Taylor’s opinions on areas of difference.
SECOND SUB-CATEGORY OF POWER QUESTIONS (S.102)
194 It is convenient for me to now discuss the s 102 question.
(a) General
195 Section 105(4) of the Act prohibits me from directing an amendment that is not allowable under s 102. The onus lies on Branhaven to satisfy me that the amendments sought are allowable under s 102. As examination of the 253 Application was requested on 7 December 2010, the form of s 102 before the amendments made by the 2012 Amending Act applies. Further, given that the complete specification has been accepted, s 102(2) also applies.
196 The issues raised by s 102 are whether as a result of the amendments the claims would claim matter not in substance disclosed in the specification as filed (s 102(1)), and whether as a result of the amendments the claims would fall foul of s 102(2)(b), that is, non-compliance with s 40(2) or (3) being relevantly for present purposes lack of fair basis, clarity and definition.
197 Section 102(1) provides that, if by reason of the proposed amendments, the specification would claim matter not in substance disclosed in the specification as filed, the amendment is not allowable. The notion of “in substance disclosed” is similar to the test of “fair basis”. It can therefore be tested by asking whether there is a “real and reasonably clear disclosure” in the specification as filed of the new proposed amended claims (Lockwood Security Products Pty Ltd v Doric Products Pty Ltd (No 1) (2004) 217 CLR 274 at [69]). The same test applies to non-compliance with s 102(2)(b) where there is lack of fair basis. The usual tests for clarity and definition also apply to s 102(2)(b). I adopt the relevant principles that I have discussed in my principal reasons on these aspects.
198 Section 102(1), (2) and (2A) relevantly provide:
102 What amendments are not allowable?
Amendment of complete specification not allowable if amended specification would claim matter not in substance disclosed in the filed specification
(1) An amendment of a complete specification is not allowable if, as a result of the amendment, the specification would claim matter not in substance disclosed in the specification as filed.
Certain amendments of complete specification are not allowable after relevant time
(2) An amendment of a complete specification is not allowable after the relevant time if, as a result of the amendment:
(a) a claim of the specification would not in substance fall within the scope of the claims of the specification before amendment; or
(b) the specification would not comply with subsection 40(2) or (3).
Meaning of relevant time
(2A) For the purposes of subsection (2), relevant time means:
(a) in relation to an amendment proposed to a complete specification relating to a standard patent—after the specification has been accepted; or
(b) in relation to an amendment proposed to a complete specification relating to an innovation patent—after the Commissioner has made decisions under paragraphs 101E(a) and (aa) in respect of the patent.
199 I would note at this point that I do not need to consider s 102(2)(a) in detail as MLA has not disputed that the claims as proposed to be amended fall within the scope of the claims prior to amendment. If I need to say it, I am satisfied that the amendments are not disallowable under s 102(2)(a).
200 Before proceeding further, I would make some other general points.
201 First, the amendment application under s 105(1A) is a new and separate question from the appeal. Now Branhaven says that I have made findings relevant to a stipulation of the degree of statistical significance for the association of a SNP with a trait and to the degree of LD required of a limb (b) SNP with a limb (a) SNP and that MLA cannot go back and seek to undermine the substantive grounds of opposition that were decided by me in my principal reasons. But I agree with MLA that my expressions of view in my principal reasons cannot prevent proper consideration of an application to amend the claims under s 105(1A) on the evidence now before me. I did not have any amendment application nor proposed amended claims before me in the appeal. To the extent that I considered amendment, it was only to suggest potential amendments that might overcome the problems with the claims. I agree with MLA that my comments were not made in the context of an application to amend claims and a consideration of whether any amended claims would comply with s 102. I had not made any relevant factual findings in relation to the amendments now proposed. The proper disposition of the amendment application requires an analysis of the claims as they stand now, the proposed amended claims and a consideration of the matter in substance disclosed in the specification. It also requires a fresh consideration of whether the claims will contravene s 40 as a result of the amendment.
202 Second, I agree with MLA that it is not sufficient in and of itself for Branhaven’s success on the amendment application, as Branhaven contends, that the effect of the amendments is to narrow the scope of the matter that is claimed to a subset of what was already disclosed in the 253 Application. Such an approach was rejected by the Full Court in AstraZeneca AB v Apotex Pty Ltd (2014) 226 FCR 324 in relation to section 114(1) at [244]:
A very general description of an invention in a specification before amendment might not contain a real and reasonably clear disclosure of more specific embodiments of the invention subsequently disclosed and claimed after amendment. … Whether or not there is a real and reasonably clear disclosure in the specification before amendment of what is claimed in such circumstances is the question that arises in this case.
203 I will address the substance of this in a moment.
(b) The association feature
204 Let me first address MLA’s arguments. It says that the association feature does not comply with s 102 because it would result in the claims not being clear or not defining the invention contrary to s 102(2)(b) and ss 40(2) and (3).
MLA’s arguments
205 Now the proposed amendments introduce the word “significantly” before associated. MLA says that this renders the claim unclear, as it is unclear how this interacts with the p-value. It thereby also lacks definition. MLA says the evidence makes clear that the term is redundant in light of the requirement of a specified p-value of ≤0.05, or alternatively ≤0.01. As such, the claim, as a result of the amendment, would not be clear and succinct as required.
206 Further, MLA says that the proposed amendments introduce the requirement that each of the at least three SNPs are associated with the trait, “with the degree of statistical significance being ≤0.05”. But it is not clear if: (a) as a result of this amendment, a person skilled in the art would be required to perform a statistical test of significance or have prior knowledge of a level of statistical significance of each SNP, in order to perform the method of the claims; or (b) as a result of the amendment, a person can fall within the scope of the claim if applying a method of genetic selection using SNPs, without knowledge of whether the SNPs are associated to the required level (or using that fact as part of the method), if it is independently established that at least three of the SNPs do meet the required level of association.
207 MLA says that the evidence of Professor Taylor was that a person skilled in the art, applying a method of using SNPs to infer a trait in a bovine subject, which does not involve using SNPs on the basis of their degree of statistical significance (e.g. genomic selection), could still fall within the scope of the proposed amended claim if it was later determined that three or more of the SNPs located within genes met the requirement of a p-value ≤0.05. MLA submits that the fact that the claims, as amended, can be interpreted in this manner indicates that they have an indeterminate scope, and that they do not provide a workable standard commensurate with that required (Minnesota Mining and Manufacturing Co v Beiersdorf (Australia) Ltd (1980) 144 CLR 253 at 274).
208 Further, MLA submits that if a SNP can be found to be associated with a trait at a later time, as proposed by Professor Taylor, this would lead to the claims lacking clarity as this would provide a “moving target”, which is determined independent of the person performing the method of inferring a trait. Professor Visscher gave evidence that the p-value and sample size are often a “moving target” in association studies, at least in human studies, as associations initially reported are commonly later invalidated. It says that the evidence shows that in the application of a statistical test to determine a p-value, there are multiple factors that influence the determination of a p-value. These include the number of animals used (i.e. the sample size) and the statistical model used, although the evidence was that this variable would fall away given the same pedigree.
209 MLA says that the evidence establishes that neither the specification nor the claims dictate the number of animals to use and that this would not have been known in 2002. And nor do the claims require a specific statistical model to test the association. Rather, this is left to those skilled in the art who may choose different models.
210 MLA says that the result of this is that the determination of which actions fall within the scope of the claims is subject to a series of decisions and selections. As such, a person skilled in the art cannot satisfactorily determine if they will fall within the scope of the claims.
211 MLA says that its position is supported by the evidence of Professor Taylor, when he conceded that even if he did perform an association study, and identified a p-value of 12% (P=0.12) based upon a sample of 1,000 Charolais animals, he would still “be nervous” that he might “fall within the scope of ... [the] claim” and he “would avoid using that SNP, even though [he] was not yet absolutely sure whether or not it fell under the scope of this claim”. Furthermore, he agreed that another person, in assessing whether the SNPs met the required p-value, could arrive at a different result if they were to employ a different sample size or statistical test.
212 MLA says that there is no guidance in the specification or the claims regarding the calculation of the p-value of the association between a SNP and a trait. As I have said, its position is that the p-value will be different depending on a number of factors including the population of cattle tested including the number and breed of cattle. Thus the p-value is not clear because it does not provide a workable standard. That is, a person skilled in the relevant art will not know whether applying the claimed method to a particular population would infringe the claims. It thereby also lacks definition.
Analysis
213 I reject MLA’s s 102 arguments on this aspect of the case. Let me first deal with the question of statistical significance.
214 The amendments stipulate a degree of statistical significance being a p-value of ≤0.05, alternatively a p-value of ≤0.01. Claims 1 to 14 specify the former value. Claims 15 to 28 specify the latter value.
215 Now as to whether this is in substance disclosed, it is no longer in dispute that the specification provides support for a p-value of either ≤0.05 or ≤0.01. Nevertheless I should say something on this aspect.
216 In my principal reasons, I indicated that usually and appropriately the level of statistical significance would be stipulated at a p-value of ≤0.05 but went on to indicate that subject to hearing further from counsel the level of statistical significance in this case should desirably be at ≤0.01 (at [266]). Nevertheless I am prepared to accept that the proposed amendments stipulating a p-value of ≤0.05 would address my concerns.
217 Now a p-value of ≤0.01 was used by the inventors to identify the 2,510 specified SNPs as being associated with traits in Example 3 of the 253 Application. But whilst that was the particular degree of association employed in the examples to identify those SNPs, it does not follow that the concept of “association” in the claims must be so limited.
218 In contrast to Example 3, [0051] provides a more general definition of the concept of association between a SNP and a trait, as previously extracted: “A SNP is associated with a trait when at least one nucleotide occurrence of the SNP occurs more frequently in subjects with a certain characteristic of the trait in a statistically significant manner, for example with greater than 80%, 85%, 90%, 95%, 99% confidence”. Although expressed in terms of confidence levels rather than p-values, this supports a broader definition of association between a SNP and a trait.
219 Similarly, [0080] of the 253 Application, noted in my reasons at [301], refers to using known statistical methods to identify relationships between SNPs and traits on the basis of “an association … with at least 80%, 85%, 90%, 95%, 99% confidence, or alternatively a probability of insignificance less than 0.05”. That language contemplates a p-value of at least 0.05 in this context.
220 Further, although the inventors chose to use the more stringent per test level of p <0.01 in both Examples 2 and 3, these passages in the 253 Application also indicate that a lower level of stringency can be used, including p-values of 0.05, 0.1 or 0.2.
221 Let me move to the clarity question.
222 MLA contends that the inclusion of a value of statistical significance of either p ≤0.05 or of p ≤0.01 renders the claims unclear.
223 Now p-values are well known and well established statistical measures, having been utilised inter-alia by statisticians in the field of genetics since at least the 1900s. And the fact that p-values may vary depending on a number of factors including the nature and size of the population of the cattle tested does not render the scope of the claims unclear.
224 The language of a claim that uses terms requiring assessment according to circumstances does not necessarily mean that the impugned claim lacks clarity. Further, relative expressions which require the exercise of judgment can be used, but they must be understood in a practical common-sense manner. Moreover, a patent applicant does not have to express its claim with a precision that implies an arbitrary restriction on the inherent variability of a feature which is part of the invention.
225 Further, in the present case the skilled addressee would understand that a sufficiently large sample size is necessary in any population genetic method to minimise sampling effects and to provide sufficient power to reject the null hypothesis when it is not true.
226 Further, the issue of false positives does not present a practical issue. A validation study would routinely be done in any case where the population of interest was genetically different to the research population; I have already said in my principal reasons that validation studies were routine (see [878] and [910]). The SNPs that were most predictive in the population of interest would be chosen as a result of the validation. The validation study would determine which of the SNPs were truly trait associated or were false positives. And any false positives would not be used.
227 In my view, the specification of the degree of association required provides a workable standard suitable for its intended use and capable of objective assessment for a person skilled in the relevant art to determine if a SNP that they were using was sufficiently “associated” with a trait.
228 Further, no evidence or argument advanced on the amendment application has thrown doubt on any of the following findings in my principal reasons:
(a) The claims “need to be amended to stipulate the degree of statistical significance required in order to establish the required ‘association’”, and “usually and appropriately that would be stipulated at a p-value of equal to or less than 0.05, but in this case desirably at equal to or less than 0.01”. (at [266])
(b) “[I]t is apparent that the claim as drafted includes any SNP covered by the claim that has any degree of association with any trait.” (at [297])
(c) The reference to “association” in the claims “should have been so expressed with some stipulation for the degree of statistical significance” and “[s]ome objective stipulation of statistical significance and the appropriate degree is necessary”. (at [302])
(d) “[I]f the amendments that I have suggested be made concerning ‘associated’ and LD are made, then I agree with Branhaven that the claims of the 253 Application include (or will include), as limiting and characterising features, requirements that ensure the utility of the invention.” (at [877])
(e) To address issues of lack of clarity and definition, the claims “require amendment to: (a) define ‘associated’ in terms of statistical significance at the p value of equal to or less than 0.01 (or such other measure as I decide after hearing from counsel further); [and] (b) require each of the 3 SNPs to satisfy that level of statistical significance (to be discussed further with counsel)”. (at [944(a) & (b)])
229 These findings were made in light of detailed expert evidence, and following a debate about the clarity of the claims. They contemplate that the stipulation of the degree of statistical significance of the association by reference to an appropriate p-value was required in order to remove an issue of lack of clarity in the claims, and that the claims would be clear in this respect if such an amendment were made.
230 Nothing that MLA has put before me on the amendment application suggests that I should now find that the introduction of the very stipulation that I referred to would result in, rather than remove, a lack of clarity of the claims. The same goes for the suggestion that this aspect of the proposed amendments would introduce a lack of definition in the claims.
231 Further, the following evidence does not support MLA’s grounds of objection on this aspect, as Branhaven correctly contended:
(a) As at December 2002, p-values were well-known and well-established statistical measures, including in the field of genetics. P-values of 0.01 or 0.05 were used in statistical methods employed in cattle before December 2002.
(b) Further, the statistical significance of the association is to be assessed in the population in which the person employing the method wishes to deploy the method. There is no basis in the wording of the claim for the use of some other reference population.
(c) Further, the skilled person would determine a sample size in the population of interest that would be relevant to determining trait-associated SNPs with a meaningful application within the population of interest. And a sufficiently large sample size is necessary in any population genetic method to minimise sampling effects and provide sufficient power to reject the null hypothesis when it is not true. The skilled person would understand that the association would need to be tested in a reasonably large sample size. And this was all within the skill of the calling.
(d) Further, the issue of false positives does not present a practical issue. A validation study would routinely be conducted in order to determine which of the SNPs were truly trait-associated or false positives. As I have already said, the latter would not be used.
(e) Further, Professor Visscher suggested that the level of association detected would depend on various matters such as the study design, sample size, correction for multiple testing error, size of population, and breed of population. But these are features of the particular way in which the person employing the method of identifying a trait chooses to do so. Further, Professor Taylor’s evidence made it clear that a skilled person would readily be able to take steps to avoid falling within the claims had he or she wished to do so, and that it was within the skill of a quantitative geneticist to select the appropriate parameters and study design for such testing.
232 Let me deal with another topic under this heading concerning adding “significantly” before “associated”. In my view this does not inject any lack of clarity.
233 The word “significantly” identifies that the association between the SNP and a trait is one which is to be assessed in terms of statistical significance, and the claim(s) as proposed to be amended goes on to specify the required degree of that statistical significance by reference to a p-value. In context, this is clear. There is no ambiguity when the claim is read as a whole. Moreover, neither expert suggested any substantial lack of clarity of this kind. In the concurrent evidence session, Professor Visscher suggested that “significantly” was redundant. But this did not mean that the claim was unclear. He understood what the word referred to: the statistical significance later stated in the claim. Professor Taylor also understood the claim. In any event, the word is not redundant. It is included to provide an antecedent to the reference “the degree of statistical significance”.
234 Finally, before passing to the next topic it is appropriate that I note that the proposed amendments make it clear that each of the at least 3 SNPs must satisfy the level of statistical significance. This requirement results in a more robust method for identifying and selecting cattle with superior genetic potential for desirable characteristics. In relation to my comment at [362] of my principal reasons that the limb (b) SNP would also need to satisfy the “association” integer, as a result of the amendments the requirement for a specified statistical level of association applies to both limb (a) and limb (b) SNPs.
(c) The linkage disequilibrium (LD) feature
235 Let me now discuss the second main question concerning s 102 dealing with LD.
MLA’s arguments
236 MLA says that the specification does not provide any direct measure or quantification of LD using any metric. It says that the only context in which LD is mentioned is in Example 3, the purpose of which was to perform “an experiment to determine the disequilibrium distance in cattle” (see at [0199]). It says that this is supported by Professor Taylor, who considered that the objective of Tables 3 and 4 of Example 3 is to determine the range over which disequilibrium extends. According to MLA, at most, LD is assumed by the inventors in this example. It says that LD is not measured in the specification by any specific metric, nor is any degree or metric for measuring LD discussed, mentioned or suggested in the specification. I noted in my principal reasons at [356] to [357] that the specification “explains that SNPs within +/- about 500,000 nucleotides of the specified SNPs are expected to be associated with the same traits as the specified SNPs. Such an “expectation” avoids or negates the need for SNPs within that region to be shown to be in LD with a specified SNP. Indeed, depending on the parameters by which LD is assessed, including population size, all SNPs in the genome can have some degree of LD with each other SNP…".
237 But MLA says that in some respects the specification indicates the following:
(a) [0199] on page 62 (last three lines) of the specification indicates that the degree of LD is variable and depends on a number of different factors.
(b) Example 3 uses distance (i.e. 500 kb either side of a specified SNP) as a proxy for LD (see [0201] and [0208] of the specification).
238 Indeed, Professor Visscher said that if an r2 value were used to indicate LD in the same way as distance is used in Example 3, the value would be expected to be 0.99 or 1.
239 Further, MLA says that the metric included by way of the proposed amendment, namely r2, is not disclosed in the body of the specification, or the claims prior to amendment, and therefore there is no basis at all in the specification for measuring LD using r2, or any other metric. Further, MLA says that a person skilled in the art would not have been led to add the r2 metric into Example 3. Further, MLA says that in addition to the fact that the specification discloses distance as a measure for LD, and therefore teaches away from any other metric to measure LD, the evidence establishes that in 2002 there was no consensus as to the appropriate metric to estimate LD in 2002. A person skilled in the art could use many different metrics including D, D’, r2 or r.
240 Further, MLA says that the only genome wide study performed in cattle prior to 2002 used the D’ metric. MLA contends that the D’ metric was still the primary metric used in relevant publications after 2002, and up to at least 2007. Accordingly, MLA contends that a person skilled in the art, having regard to what was known in the art at the time of filing the 253 Application would not have concluded that LD as described in Example 3, would have inferred measurement by r2. Therefore, according to MLA, there is no implicit disclosure in the specification of r2. As such, so MLA submitted, the amendment to include such a feature into the claim, will result in the introduction and claiming of matter not “in substance disclosed” in the specification and accordingly will not comply with s 102. It is new and original material. Further, it says that a conclusion that a feature can be added to a claim by way of merely being known as part of common general knowledge would lead to the common general knowledge serving as a reservoir of features to be added to a claim in any desired combination.
241 Let me now deal with another dimension to MLA’s arguments.
242 The amendments proposed to the claims further attempt to introduce an r2 value having a threshold ≥0.7. But MLA contends that any introduction of a threshold is purely subjective, and has no basis in the disclosure of the specification nor any objective basis in the common general knowledge. Further, Professor Taylor conceded that there was no disclosure of any particular r2 value as a minimum or of an r2 value of 0.7 or 0.8 in the specification.
243 It is also said that Professor Taylor further conceded that the recommended amendment of an r2 ≥0.7 is based on his opinion of what the majority of geneticists would consider to be strong or high LD. He interpreted the claims in the specification to require a high level of LD between limb (a) and limb (b) SNPs. Alternatively, the claims could be interpreted to include a moderate level of LD. I would note that Professor Taylor said that most quantitative geneticists would consider r2 greater than 0.7 to be strong LD. He did accept, however, that views might reasonably differ as to what “moderate” might mean or what “high” or “strong” might mean, although most quantitative geneticists would consider r2 greater than 0.7 to be “strong” or “high”.
244 MLA says accordingly that the evidence establishes that the distinction of the bounds defining low, medium and high LD were subjective and that others skilled in that art would have their own interpretation of what those words would mean, and they in fact had been used differently in publications of which Professor Taylor was an author.
245 Accordingly, MLA submits that the proposed amendments to the claims to introduce a threshold based on an arbitrary, subjective, and non-scientifically-based limit would introduce subject matter to the claim with no basis on matter disclosed in the specification, nor with any basis on that which is known and agreed upon in the art. Consequently, it says that this would lead to the introduction of matter not “in substance disclosed” in the specification.
246 Let me discuss a yet further dimension to MLA’s concerns.
247 MLA submits that the evidence establishes that the introduction of an r2 level to a specified threshold results in the amended claims lacking clarity on the basis that: (a) LD between the two relevant limb (a) and limb (b) SNPs will differ between populations; (b) the accuracy of predicting LD depends on sample size; (c) it is unclear if the threshold value for LD applies to the “true value” of LD, or the “estimate value” of LD; and (d) if the threshold applies to estimate value, then it is unclear if a statistical test needs to be applied to the threshold, what p-value would be used to test the estimate value and what sample size would be needed to test the estimate value.
248 Further, according to MLA, Professor Taylor asserted that the r2 value would need to be tested by use of an unspecified statistical test, to an unspecified p-value, on an unspecified sample size. But I think that one needs to be more precise at this point in terms of the evidence given by Professor Taylor. Professor Taylor gave evidence that although these parameters were unspecified he would understand the r2 value discussed in the specification to represent an “unknown population parameter”, which is not the same as a sample estimate. Professor Taylor gave evidence that in applying the method he would first assemble an appropriate sample, genotype the SNPs of interest, estimate the r2 value between pairs of SNPs and then apply a Fisher transformation to test the hypothesis for the relevant r2 value of the population by using a p-value of <0.05 in order to reject the null hypothesis. All standard statistical work according to Professor Taylor for a person skilled in the art with common general knowledge. I agree. But according to MLA, Professor Visscher’s evidence suggests that this is not a standard step in the art, and that he has never performed such a statistical test on an r2 value. Further, Professor Visscher said that such a requirement as put forth by Professor Taylor was “confusing”. But when further questioned by counsel for Branhaven he agreed that this was a method which could be used. I have already said that I prefer Professor Taylor’s evidence given that he had the edge on practical industry experience.
249 MLA also says that the requirement to assess the significance of the r2 value with a non-prescribed p-value would lead to the exact problem noted by me in my principal reasons at [266] that:
If statistical significance is not stipulated, there is in my view no adequate measure for or objectivity enshrined in the concept “association” or its cognate verbial forms. Subjectivity and imprecision would rule the day. In my view, absent a proper stipulation for statistical significance, the claim lacks clarity and the invention has not been properly defined.
250 Further, MLA says that even accepting that a person skilled in the art would know to perform a statistical test on an r2 estimate of LD, and would know what p-value to use, they would still not know what sample size to use. This would lead to a lack of clarity and would not provide a workable standard to a person skilled in the art.
251 It says that the lack of clarity is exemplified by the hypothetical situation put forth by Professor Taylor, where he explained that even if he obtained an estimate of 0.65, that would not tell him that the true value of r2 is not greater than 0.7, which Professor Taylor said would require statistical testing.
Analysis
252 Let me begin with and repeat what I said in my principal reasons:
(a) “[I]t will be necessary to amend the claim so that limb (b) makes it plain that relevant LD is stipulated. Otherwise the claim lacks clarity and does not properly define the invention.” (at [265])
(b) “LD should be required and stipulated in limb (b) to avoid problems dealing with a lack of clarity and a failure to define the invention.” (at [267])
(c) “LD is determined by measuring the frequency at which the presence of two markers (such as SNPs) are found together within individuals in a population.” (at [359])
(d) “LD is not an all or nothing concept, but a matter of degree. LD is measured between 0 and 1 … If the degree of LD between a specified SNP and a non-specified SNP is very low, the non-specified SNP would not be expected to be a useful surrogate for the specified SNP.” (at [359])
(e) “[L]imb (b) fails for lack of clarity absent amendment. First, the LD requirement should be explicitly stated. Second, a meaningful degree of LD should be explicitly stated, so that a non-specified SNP (limb (b)) can be considered to be a useful surrogate for the specified SNP (limb (a)).” (at [362])
(f) “In order to assess LD in this context, it would not be necessary to conduct an association study, but rather to genotype the SNPs in the target population and assess whether they appear to be associated with each other in a non-random fashion across the population. I agree with Branhaven that such an approach was routine and well within the skill of the calling.” (at [374])
(g) “[I]f the amendments that I have suggested be made concerning “associated” and LD are made, then I agree with Branhaven that the claims of the 253 Application include (or will include), as limiting and characterising features, requirements that ensure the utility of the invention.” (at [877])
(h) To address issues of lack of clarity and definition, the claims “require amendment to: … require the limb (b) SNP to be in LD with the relevant limb (a) SNP and to the requisite degree (to be discussed further with counsel).” (at [944(c)])
253 Clearly, my observations based upon the evidence then adduced contemplated, as Branhaven points out, that the stipulation of an explicit requirement of LD between a limb (b) SNP and the relevant limb (a) SNP, and the specification of a requisite degree of LD by reference to an objective measure, were required in order to remove an issue of lack of clarity in the claims, and that the claims would be clear in this respect if such an amendment were made.
254 As should be apparent, what remained to be considered was both the methodological measure to be used for LD and the degree of that measure to be stipulated.
255 Now the specification discloses a requirement for LD between a limb (b) SNP and a limb (a) SNP. At the least this requirement was implicitly disclosed though not as to degree. But it is clear that a very high or perfect LD is not being insisted upon in the 253 Application i.e. the disclosure is broader; see at [0035], [0126], [0127], [0198] to [0201], [0205] to [0208].
256 Let me now turn to discussing the evidence in more detail.
257 In terms of the evidence adduced before me at trial and on the amendment application, the following should be observed.
258 First, the r2 measure was a standard statistical measure which was known in December 2002, including as a measure of LD. Indeed, Professor Visscher accepted that it was a known measure.
259 Second, a skilled person reading the 253 Application in December 2002 with the common general knowledge would have known what r2 was and might have used it as a metric to measure LD. Professor Visscher also accepted this.
260 Third, in a 2005 paper of which he was an author, Professor Visscher used a measure of “say, r2 > 0.3”, which was described as a “useful” LD (Uimari P et al., “Genome-Wide Linkage Disequilibrium from 100,000 SNPs in the East Finland Founder Population” 8(3) (2005) Twin Research and Human Genetics 185).
261 Fourth, there is an explicit reference to “measures” of LD in the 253 Application. It is apparent that r2 was one such measure before December 2002. Professor Visscher so accepted this.
262 Fifth, Professor Visscher understood from reading the claim what an r2 value of greater than or equal to 0.7 was, and accepted that this was something he would have been capable of measuring as at December 2002.
263 Sixth, Professor Taylor explained how he would have gone about measuring an r2 value of LD, using standard statistical approaches. And the fact that the claim does not specify a particular sample size or details of the statistical test is not to the point. These are parameters routinely chosen by the skilled quantitative geneticist in order to derive an estimate of such a value.
264 Seventh, Professor Visscher accepted that such a statistical approach could be adopted, that an estimate of r2 with confidence intervals could be derived using such an approach, and that if a person wanted to increase the precision of the estimate a larger sample size could be used.
265 Eighth, Professor Visscher also accepted that one would not need LD of between 0.99 and 1.0 in order to identify a limb (b) SNP that was also associated with the same trait as a limb (a) SNP, and that insistence upon such a high degree of LD could result in excluding a limb (b) SNP that was more strongly associated with a relevant trait(s) than the corresponding limb (a) SNP. Moreover, Professor Visscher accepted that the specification in relevant passages did not insist upon an LD as high as between 0.99 and 1.0.
266 Generally, as is apparent, significant aspects of the evidence of Professor Visscher supported Branhaven’s case. Let me now turn to address MLA’s s 102 points directly.
267 MLA’s first point is that there has not been in substance disclosure and, relatedly, that there is no fair basis.
268 MLA has contended that an amendment that introduces an entirely new integer or new and original material is not allowable on the basis that it is not “in substance disclosed”. It contends that the 253 Application did not disclose a particular measure of LD, being r2, and at a particular value, being ≥0.7 or ≥0.8. Therefore, the introduction of such a metric and at such a value was not “in substance disclosed”. Let me deal with this submission at a number of levels.
269 First, I did not find the UK cases to which my attention was drawn to be that helpful to my consideration of what was or was not “in substance disclosed” in the 253 Application. Cases such as AMP Inc v Hellerman Ltd [1962] RPC 55 at 72, Ethyl Corporation’s Patent [1972] RPC 169 at 192 to 195 and Shionogi & Company Ltd’s Application [1967] RPC 623 at 626 and 627 turn entirely on the particular specifications and particular amendments sought. Further, what was sought to be added in Shionogi (“new and original material so as concerns…the preparation of the starting material and…the separation technique…”) is a far cry from what I am considering. In my case what is being considered, on the hypothesis that LD has been disclosed, is the measurement and narrowing degree thereof. It is hardly a new integer. Further, AMP is well away from what I am considering. That was dealing with the situation where “[t]he tool with a stop was disclosed as well as the tool without a stop. The patentee disclaims the tool without a stop and confines himself to the tool with a stop”. Lord Denning saw no problem as the amendment had not sought to add “an entirely new integer”. Further, Ethyl Corporation’s Patent really does not assist MLA either although it is a little closer. The amendment proposed to claim 14 introduced a restriction into that claim stipulating the ratio of a hydrocarbon to tetramethyllead in an antiknock composition with the “hydrocarbon being present in amounts from 20 to 80 weight per cent of the tetramethyllead”. The amendment was allowed because the original specification contained examples of compositions within the amended added range. But it is a leap to extrapolate from that case that non-reference to a metric or its degree in relation to an integer which serves a different significance in the case before me can be said to be not “in substance disclosed”. MLA said that the UK cases demonstrate that “the test has been applied strictly”. I am not sure where the word “strictly” comes from other than being MLA’s characterisation. But what these cases demonstrate is that entirely new integers and new and original additions to the invention are not permitted. But that is not the case before me.
270 Second, the present amendments sought by Branhaven are narrowing amendments. If it is accepted that LD has been disclosed, as it must be, the amendments seek to clarify and narrow how it is to be measured and the degree thereof. Indeed I have already found in substance that there is fair basis with respect thereto.
271 Third, if it be correct to say, as I think it is, that there is a close relationship between the test for in substance disclosure and the test for fair basis, then the observations of Yates J in DSI Australia (Holdings) Pty Ltd v Garford Pty Ltd (2013) 100 IPR 19 at [240] are useful:
In the present case, Garford submitted that the notional claims propounded by the DSI parties were not fairly based on the matter described in the IRF application. I am of the view that the notional claims, if they were to be claims of the IRF application, would be fairly based on the matter described in the specification of that application for the purposes of s 40(3) of the Act. I am satisfied that there is real and reasonably clear disclosure in the body of the specification of the invention that is notionally claimed. Importantly, in this connection, the inquiry as to fair basis is directed to the question of claim width: see, for example, Olin Corporation at CLR 240; ALR 152; IPR 200. A claim may be fairly based for the purposes of s 40(3) of the Act where it adds a feature to a combination otherwise described in the specification and, by that addition, limits the described invention, as a matter of definition, to a more restrictive form than that to which the patentee might otherwise be entitled. In short, a claim may be fairly based for the purposes of s 40(3) of the Act even when all the characteristics by which the invention is defined in the claim are not described in the body of the specification itself, provided those characteristics are truly limiting ones in the sense that I have described.
272 It is apparent that there may be no need for explicit disclosure in the specification of truly limiting features; I am not here dealing with the “very general description of the invention” case of the type discussed by the Full Court in AstraZeneca, and in any event that Court expressed itself in terms of “might not contain” (my emphasis). In the present case the measure of LD and its degree which are being added are truly limiting features and matters of detail to the LD aspect already disclosed.
273 Fourth, MLA as I understood its argument said that you could not just pluck out anything from common general knowledge and add it by way of amendment, in other words r2 and its degree. Of course put so glibly that proposition is correct. But that is not what Branhaven is doing. Branhaven is narrowing its claims consistently with well understood concepts forming part of the common general knowledge as at the priority date. It is not picking an “entirely new integer” for the invention from common general knowledge.
274 Fifth, there is no dispute that it was well known at the priority date that the degree of LD varied. This understanding is consistent with the specification which, as I have said, describes at [0199]: “The degree of LD varies considerably throughout the genome and is function of time, recombination events, mutation rate and population structure.”
275 Sixth, the specification indicates that the usefulness of the limb (b) SNPs in identifying traits arises from them being in LD with the limb (a) SNP (at [0035]). In particular, the specification specifies (at [0126]) that:
In another embodiment of the invention, a method is provided for identifying SNPs that are associated with a trait by using the associated SNPs disclosed herein. The method is based on the fact that other markers in close proximity to the associated SNP marker will also associate with the trait because markers in linkage disequilibrium with the associated SNP marker will also be in linkage disequilibrium with the gene(s) influencing the trait. SNPs in linkage disequilibrium can be used in lieu of determining a SNP or mutation to predict the presence or absence phenotypic trait or a contributor to a phenotypic trait. Accordingly, in certain embodiments, the present invention provides a method for identifying a SNP associated with a trait, that includes identifying a test SNP that is in linkage disequilibrium with a [limb (a)] SNP.
276 In my view in its proper context the skilled addressee would understand that the reference to LD between the limb (a) and (b) SNPs in the specification is, practically, a reference to a need for “high” or “strong” LD. An r2 value of 0.7 and above is consistent with this.
277 Seventh, MLA’s principal contention is based on the fact that the specification does not explicitly identify any method for measuring LD including the r2 measure. But this ignores that the specification is not to be read in the abstract but in the light of the common general knowledge. The Court is to place itself “in the position of some person acquainted with the surrounding circumstances as to the state of [the] art and manufacture at the time” (Kimberley-Clark Australia Pty Ltd v Arico Trading International Pty Ltd (2001) 207 CLR 1 at [24]).
278 In that context, all that is being done in the proposed amendments is to use a common general knowledge measure of LD to provide an explicit definition of a feature of the invention that is already disclosed (implicitly and explicitly) in the description of the 253 Application: a requirement for LD between a limb (b) SNP and a limb (a) SNP. This involves merely clarifying or claiming a subset of what is already in substance disclosed, and does not result in any lack of fair basis.
279 Eighth, the evidence shows that stipulation of a “high” degree of LD as proposed is consistent with the disclosure in the 253 Application in that the usefulness of the limb (b) SNPs arises from them being in LD with a limb (a) SNP. A higher degree of LD directs the method to more useful limb (b) SNPs.
280 In that context, in my view the skilled addressee would understand that the reference to LD between the limb (a) and (b) SNPs in the specification to be or at least to include a reference to a need for “high” or “strong” LD. An r2 value of 0.7 and above is consistent with this.
281 Further, it is not to the point that no measurement of LD by the r2 measure is explicitly disclosed in the description. The concept of LD between a limb (b) SNP and a limb (a) SNP is clearly disclosed as a basis for the limb (b) aspect of the invention, as was the fact that LD is a relative concept that can vary. Moreover, as at the priority date, r2 was a common general knowledge measure of LD. Now other measures of LD were available in December 2002 and could have been used. But the r2 measure met that description. Further, the use by Professor Taylor and others of terms such as “high”, “moderate” and “low” LD in different contexts in the literature is not really to the point. The proposed amendments specify a degree of LD in terms of a specific r2 value which is objective and clear, and would have been measurable by the skilled person in December 2002.
282 Let me now deal with MLA’s second point concerning a lack of clarity and a failure to define the invention.
283 Now Professor Visscher characterised the specified r2 values of 0.7 and 0.8 as “arbitrary”. But this is not a proper basis to find that the proposed claims lack clarity. The question is whether the integer in question is clear, in the sense of it being readily capable of being understood and applied by the skilled person. In my view an r2 value of equal to or greater than 0.7 or 0.8 meets that description.
284 Of course an r2 value of equal to or greater than 0.7 or 0.8 is in one sense “arbitrary”, in that different quantitative geneticists might choose to define slightly different values as implying strong LD. But in that sense, any stipulation of a parameter of this kind in a patent claim could be said to be “arbitrary”. On any view it is a particular but readily comprehensible limitation on the claim. Indeed, the values specified in the claims are no more “arbitrary” than the 0.99/1.0 value specified by Professor Visscher. I agree with Branhaven that the specified values in the proposed amendments provide a clear and workable standard that would be understood by the skilled addressee and readily measured.
285 Further, the proposed degree of LD is high and therefore clearly sufficient for the limb (b) SNP to be used in lieu of the specified limb (a) SNP. The proposed amendment is also generally consistent with the passages in the 253 Application at [0035] and [0126]. Further, as I have said, the amendments also require the limb (b) SNP to be associated with the trait at a high level of statistical significance.
286 Now as Professor Taylor said, which I accept, Professor Visscher’s requirement for an r2 value of 0.99 or 1.0 in effect requires that the limb (b) SNPs have genotypes in the population that can be coded to be identical or almost identical to the genotypes of the limb (a) SNP. But the 253 Application does not require that the limb (b) SNP be perfectly or nearly perfectly predictive of the limb (a) SNP. The purpose of the method described in the 253 Application is to identify and select desirable traits in cattle through associations between SNPs and the trait, not to identify associations between candidate diagnostic SNPs.
287 This is apparent from the specification at [0126] and Example 3 which identifies the usefulness of other SNPs within the region (the limb (b) SNPs) to identify and select desirable traits through association, not to predict the genotype of the limb (a) SNP. This is also apparent from the discussion of Example 3 which indicates that the additional SNPs were identified by reason of their association with the trait (and therefore their usefulness in identifying traits of interest) and not by reason of genotypic identity with the limb (a) SNP. I accept Professor Taylor’s evidence to this effect.
288 Moreover, because the limb (a) SNP is in strong LD with a causal genetic variant, that is, causes trait variation, and because the limb (b) SNP is also in strong LD with the same causal variant, in my view strong, but not perfect or near perfect, LD between the limb (a) and limb (b) SNPs would be expected.
289 Further, Professor Visscher’s requirement for an r2 value of 0.99 or 1 also fails to recognise that there may well be limb (b) SNPs which are not in perfect or near perfect LD with the limb (a) SNP but that have a stronger association with the trait than the limb (a) SNP. As Professor Taylor said, such limb (b) SNPs would clearly be very useful in identifying trait(s) through association even though they are not genotypically identical or nearly identical to the limb (a) SNP.
290 Let me turn to another matter. I agree with Branhaven that the fact that the r2 value would depend on the sample size does not introduce a lack of clarity.
291 As with the determination of the degree of association between the SNP and a trait, the precision of estimation of r2 values will depend on sample (population) size. However, the hypothesis that r2 is greater or less than certain threshold values such as 0.7 can be statistically tested using the data that were generated to estimate the r2 value. Furthermore, it would be understood by the skilled addressee that a reasonable sample size was required in order for the null hypothesis to be rejected when it was not true.
292 Further, as Professor Taylor said, there were many association studies, albeit not genome-wide, between genetic markers and traits conducted before the priority date, both in relation to cattle and other species including humans. In all of these studies, the methodology including the sample size was designed so that meaningful and useful associations could be detected.
293 I reject MLA’s s 102 arguments on lack of clarity and a failure to define the invention concerning r2 and the threshold values for measuring LD.
(d) Is an association study required?
294 During the course of the hearing on the amendment application I raised with the parties whether performing the method in claim 1 and associated claims requires that a determination of association actually be carried out, or whether it simply requires that the SNPs in fact have the requisite degree of association without the person performing the method necessarily having to determine that association.
295 Now I would of course note that if performing the method in the claims required a determination of association to be carried out between the SNP and a trait prior to the proposed amendments, the proposed amendments would not introduce that requirement per se, but rather add a stipulation of the degree of association to address the issues of clarity, definition and utility found by me (see at [944(a), (b), (c)], [945]).
296 Now Branhaven submitted that MLA has never contended for a construction of the claim(s) that requires association to be determined as part of performing the method. It did not advance such a construction on the appeal. Rather, its case was based on the assumption that no assessment of association was required as part of the claimed method.
297 Branhaven submits that the claim(s) does not require that an association study be conducted or, more precisely, that the degree of association of a SNP with a trait be determined and characterised in terms of its statistical significance, in order to practise the method. Branhaven accepts that although that is one way in which a person could practise the method, the claim(s) does not require it. If a person wishes to assess whether they fall within the claim(s), they can test for the presence of that integer by determining the association of a SNP with a trait, but they do not need to do so in order to practise the method.
298 Branhaven also says that such a construction of the claim(s) does not render the claim(s) unclear. To the contrary, the requirement in the claim(s), as proposed to be amended, that a SNP be associated with a trait to the stipulated degree of statistical significance provides the objective measure of association which I found was required in order for the claim(s) to be clear. According to Branhaven, what matters is the degree of association that exists in fact, not whether the person performing the method happens to determine it and characterise it in terms of it involving a p-value of equal to or less than 0.05 or 0.01.
299 Branhaven contends that in this regard, the requirement for association and the degree of association required by the proposed amendments should be understood in the same way as the requirement for LD and the degree of LD between the limb (b) and limb (a) SNPs which are to be introduced by the amendments. It says that it is not suggested that the method requires that a person actually determine the degree of LD between the limb (b) and limb (a) SNPs in order to work the method. Rather, both features are inherent requirements. All that is required is that they are present. The skilled person can readily test for them if he wishes to do so.
300 Branhaven says that this construction of the claim(s) is supported by the plain words of the claim(s), which do not include any statement to the effect that an association study must be conducted as part of performing the claimed method. It is also consistent with the description of the invention in the 253 Application. The specification indicates that the method is directed to the utilisation of the fact of the association of a SNP with a trait, not the collection of data demonstrating or confirming that association.
301 So, [0051] of the specification states that: “… in certain aspects, the methods include identifying whether the nucleotide occurrence is a bovine SNP allele identified herein as associated with a trait” (emphasis added). In other words, the practising of the claimed method in this aspect requires identifying whether a SNP is one of those identified in the specification as being associated with a trait. For the purpose of this aspect of the claimed method, the identification of the association between that SNP and the trait is presented in the 253 Application, not determined by the skilled person.
302 Similarly, Branhaven says that [0141] and [0152] of the specification discuss the generation of high density SNP maps and SNP chips that can be utilised in methods of the invention to infer traits. In particular, [0152] states: “… as indicated above the system includes a solid support or other method to which a series of oligonucleotides can be associated that are used to determine a nucleotide occurrence of a SNP for a series of bovine SNPs that are associated with a trait”. This is the SNP chip concept. The use of such a device enables a user to identify the presence of SNPs associated with a trait in a nucleic acid sample by exposing the sample to the device. The device is designed to detect the presence of SNPs in a sample, wherein those SNPs have previously been determined (e.g., by the manufacturers of the device) to be associated with a trait in a relevant population of animals.
303 Branhaven says that as with the claims, there is no statement in the description in the 253 Application that an association study is required to be conducted as part of practising the method.
304 Branhaven also submits that Professor Taylor did not consider that actually performing an association study was a necessary step in performing the claimed method. But he did give evidence to the effect that if he was aware of the claims, he would conduct a test for association in order to determine whether the SNPs had the degree of association specified by the proposed amended claims. And such work, which I accept, was routine as at December 2002. But that is a different thing from saying that it needed to be done to perform the invention.
305 Further, Branhaven says that the determination of association between a SNP and a trait is what may be termed a “pre-solution activity”. The claimed method utilises the fact of the association to infer traits, however the collection of that information including as to the degree of association does not form part of the claimed method itself.
306 Further, Branhaven says that as a practical matter, some kind of determination of association would have been made prior to or at least as part of the claimed method being performed. In the case of limb (a) SNPs, that work was done by the inventors and the results of it were presented in the 253 Application. A person could use those SNPs without the need to test for their association and to determine its degree in terms of statistical significance. Alternatively, the work may have been done by a third party, such as the manufacturer of a SNP chip. If that were not so, then the SNPs would not have been selected in the first place. SNPs are selected to be useful in inferring traits, including SNPs used on SNP chips, because they are associated with the trait.
307 Further, Branhaven says that any requirement that an association study be conducted by the person performing the method in order to fall within the scope of the claim would lead to absurd results. For example, on that construction of the claim, a person could use the limb (a) SNPs disclosed in the description, for which the association has already been determined by the inventors, to infer traits without infringing the claim. As a practical matter, there is no need to conduct any further determinations of association for those SNPs because this information is provided in the specification. Similarly, once SNPs had been characterised by other means as having an association with a particular trait, third parties would be able to use the method without infringing the claim because there would be no need for them to determine association again.
308 Further, Branhaven says that the fact that association may be determined independently of a person performing the method does not give rise to a lack of clarity. The proposition that this would involve a “moving target” does not withstand scrutiny. The assessment of the association of a SNP with a trait in a particular context is something clearly within the skill of a quantitative geneticist. I agree. That will necessarily involve selecting a reasonable sample size for the given population. Further, it is within the ordinary skill to determine a sample size from which the extrapolation can be made to a more general population. Those basic principles are not affected by whether the association is determined by the person practising the method or has been previously determined by another person. I agree.
309 Let me turn to MLA’s position. MLA points out the following passages from my principal reasons.
310 I said the following at [290] to [292]:
Branhaven says that the claim requires the existence of a statistically significant or meaningful association with the trait of interest, but does not place any precise limit on the “degree” of association required. It is said that this is appropriate given the nature of the claimed method. It contends that the incorporation of a hard limit would arbitrarily and artificially restrict the claim, and that the level of accuracy required of the method and thus the degree of association that is appropriate to put it into effect will vary from case to case. It is said that this will depend on the application to which the method is to be put including matters such as the purpose of drawing the inference, the size of the target population, the nature of the trait of interest, the number of SNPs to be employed in the method, and the like. It contends that the evidence confirmed that the skilled person would readily be able to address these matters in a particular case.
Branhaven says that it is not the case that the claim covers any degree of association between the SNPs and the trait when determined by any means. It is said that the association must be such as to allow the claimed method to be carried out according to its terms: i.e. to enable the skilled person to draw an inference about the potential for the trait to exist in the subject. It says that the accuracy required of that prediction is a matter for the skilled person in the particular case and that this is the result of a practical and common sense construction of the claim.
It also contends that there is no substance to the contention that the claim is unclear because the skilled person will not know whether the particular associations between SNPs and traits fall inside or outside the scope of the claim. It says that to so contend misunderstands the subject matter of the claim. It is said that the claim does not claim particular associations between SNPs and traits. It claims a method of identifying a trait in a bovine subject, based on the identification from a nucleic acid sample of the bovine subject of at least three SNPs associated with the trait. It is said that the skilled person will readily understand when he or she is employing such a method, and identifying SNPs for that purpose. It says that the degree of association will depend on the particular case.
(Original emphasis.)
311 I then said at [294] that I had difficulties in accepting Branhaven’s submissions.
312 At [297] I then said the following:
First, it is apparent that the claim as drafted includes any SNP covered by the claim that has any degree of association with any trait. In my view the claim lacks clarity because although a non-zero association might be determined by one study, it might be found not to have any association at all when tested using a different study. Moreover, the fact that the claim includes any degree of association also gives rise to lack of utility.
313 Now I would note that although I addressed the phrase “at least three SNPs are associated with the trait”, I did not say that this phase required that an association study be performed as part of the method of identifying a trait in a bovine subject (see at [298] to [302] that I reproduce later).
314 Further, MLA made reference to Branhaven’s submissions concerning one of the prior art documents, Meuwissen that I dealt with in my principal reasons, which MLA asserted anticipated the claim(s) and which proposition I rejected. MLA made reference to [559], [573] and [574] of my principal reasons which said:
MLA says that the only difference in approach is the statistical method used. In Meuwissen, two of the four algorithms assumed all markers potentially have a degree of association with a QTL or gene that influences a trait (Bayes A and BLUP). The other two algorithms (the least squares and Bayes B approaches), looked at each SNP individually and tested for association on a level of stringency; I would note that a least squares approach also resonates with a GWAS style approach as Professor Taylor suggested. Further, MLA says that while Meuwissen says that the least squares approach is the least effective, it does not say it does not work. In fact, so MLA contends, it did work, just not as well as the others; MLA relies upon Professor Hayes’ evidence to this effect. In any event, so MLA contends, Meuwissen discloses no such reservations about the Bayes B method, which only uses markers which are statistically significant. MLA says that the 253 Application tests each SNP individually using an algorithm that is close to the least squares approach used in Meuwissen. In any event, MLA says that the fact that the algorithms used in Meuwissen and the 253 Application were not identical is not relevant, as the claims do not require association to be determined in any particular way, and therefore encompass all methods of determining association.
…
Both Professor Goddard and Professor Hayes accepted that the BLUP and the Bayes A methods do not identify particular markers associated with the trait. Rather, such methods use all the markers in the dense marker map in the analysis. Further, Professor Goddard accepted that one way of applying Meuwissen to real animals and real markers was to use the BLUP and Bayes A method of analysis. But as I have said, these would not identify particular markers with a trait. Contrastingly, it was accepted that the other two methods, Bayes B and least squares do identify specific markers associated with a trait. Moreover, Professor Goddard and Professor Hayes also accepted that of the 4 methods, the least squares method did not work as well as the others.
Now given just that evidence alone, and my own review of Meuwissen, it follows that the direction in Meuwissen (if applied to real animals and real markers) is at least as likely to be carried out in a way by using BLUP and Bayes A that does not identify specific markers associated with a trait. But that is not what is required by the claims of the 253 Application.
315 Leveraging off these references, MLA says that it appears that I found that the claims required, as a step of the method of inferring a trait in a bovine subject, the act of identifying an association between the SNPs in question and the trait in question. So, it was not sufficient to simply conduct a BLUP or Bayes A method when inferring a trait in a bovine subject, because those methods did not involve the identification and selection of SNPs on the basis of the SNPs being identified as having a statistically significant association above a specified level of stringency such as a p-value.
316 MLA says that during the evidence of Professor Taylor on the amendment application, it emerged that there was a question as to whether the claim required a statistical test of an association to be performed as part of the method of claim 1. During closing submissions, MLA submitted that if the claimed method required both the identification of an association between the SNPs and trait, and the determination that that association was statistically significant, then the claim did not lack clarity by reason of introducing a statistical test of the association, but otherwise it did.
317 In response to a question from me on this issue, Branhaven’s senior counsel stated that its position was that claim 1 “as it presently stands, and as proposed to be amended, is to be understood not as requiring any association, study, or assessment to be conducted in order to implement the method”.
318 MLA says that it appears that I have already decided that the association integer is to be construed as requiring the conduct of an association study to be performed as part of the method of inferring a trait. It says that it also appears that I have determined that that association study must also include a statistical test of the identified association(s), rather than simply the identification of a mere “correlation”, the latter not requiring an additional test of statistical significance.
319 Alternatively, MLA says that if I have not already determined that the claims require performing a statistical test of an identified association as part of the claimed method of inferring a trait, the proposed amendment would introduce a lack of clarity in that it is not clear whether the statistical test must also be performed as part of inferring the trait or need only be an inherent feature of the SNPs in that claim, and thus determined at any time, by any person, in any population, of any size and using any statistical test.
320 Further, MLA says that the introduction of a degree of association (i.e. a p-value) into the claim introduces a new ground of lack of clarity. An association may be determined as statistically significant to the requisite degree by one study, but not in a different study.
321 Finally, MLA says that a question for me now is whether I should direct that the 253 Application proceed to grant, as proposed to be amended, in the knowledge that Branhaven asserts that the claim has a different meaning to that found by me as appears to be the case.
322 Now in my view, the claim(s) either in their present form or as proposed to be amended do not require an association study to be carried out as part of the claimed method. And if my principal reasons can be read as suggesting otherwise I take responsibility for that lack of clarity. But in my view the claim(s) do not require that the association of a SNP with a trait or the degree of that association be determined or characterised in terms of its statistical significance by the person practising the method. As Branhaven correctly submits, the question is whether the SNPs have the required association in fact.
323 Further, there is a distinction between identification of the occurrence of a SNP in a nucleic acid sample, on the one hand, and determination and characterisation of the degree of association between that SNP and a trait, on the other hand. The claims require the former, but not the latter. This is apparent from the wording of claim 1, which states that the method “compris[es] identifying in the nucleic acid sample an occurrence of at least three … SNPs”, and that those SNPs “are associated with the trait”, but does not require that that association or its degree be determined or characterised as part of the method.
324 Further, as to MLA’s references to Branhaven’s submissions concerning Meuwissen and my discussion thereof in my principal reasons, Branhaven was making the point that the claimed method requires the identification of the occurrence of particular SNPs that are in fact associated with a trait, whereas the prior art did not. Branhaven’s submissions did not contend that the claims are novel because they require the person practising the method to carry out a step of determining and characterising the association of the SNPs with the trait.
325 Moreover, in my principal reasons at [574] (reproduced above) I hope I was making a similar distinction. Meuwissen was distinguished on the basis that if practised in the real world it could be carried out in a way that did not involve the identification from a nucleic acid sample of any particular SNPs associated with a trait. In this regard, MLA’s submission elides the distinction between identifying the occurrence of SNPs that are in fact associated with the trait, and to use MLA’s words “identifying an association between the SNPs in question and the trait”. My reasoning and the claims are directed to the former, not the latter. Moreover, there were many other reasons why Meuwissen did not anticipate (see at [567] to [577]).
326 Further, MLA does not identify any passages in the specification which are said to support its construction of the claims. In my view, reference to the body of the specification supports Branhaven’s construction, which accords with the plain words of the claims. It includes, for example, the use of a “SNP chip” device, a process that involves the identification of the occurrence of SNPs that are associated with a trait, but does not require any determination or characterisation of that association.
327 In summary, if one focuses on the wording of the claims as proposed to be amended, and how those claims should be construed:
(a) the claims do not in terms require that the degree of association of a SNP with a trait be determined and characterised as part of the claimed method;
(b) the description in the specification does not impose any such requirement, and to the contrary confirms that no such step is required as part of the method;
(c) the claims are clear and readily capable of being applied by the skilled person in this form;
(d) in this regard, the association requirement should be understood in the same way as the LD requirement to be introduced into limb (b), which does not require that the skilled person actually determine the degree of LD between the limb (b) SNP and the limb (a) SNP in order to work the claimed method; and
(e) this reflects a common sense understanding of the claims, such that it would ensure that the claims could not be circumvented by a skilled person simply by using the limb (a) SNPs identified in the specification, or by using limb (b) SNPs identified by other means, without themselves characterising the association of those SNPs with the trait of interest as part of their practise of the method.
328 I also reject MLA’s alternative contentions that a lack of clarity is now apparent as it is at least unclear, in carrying out the claimed method, whether the relevant statistical test must also be performed as part of inferring the trait.
329 Finally, as to the point made again by MLA that introducing a p-value introduces a new ground of lack of clarity because an association may be determined as statistically significant to the requisite degree by one study but not in a different study, I would repeat what I said earlier regarding p-values and my discussion thereof.
330 Now MLA has sought to leverage off what I said in my principal reasons at [297]. But I think the context is all important. I was in this part of my reasons dealing with the meaning of “associated” beginning at [285]. At [295] to [303] I said the following:
Before I proceed further, let me be clear as to what is being precisely talked about here. I am concerned to consider “association” in terms of a correlation with a trait. I am not concerned at this point with the quantum of the percentage variation in a trait to be explained by a SNP(s). But as soon as one talks of correlation, one really needs to consider statistically significant correlations. After all, a correlation that has no statistical significance is useless as an objective measure or observation of anything. It is a correlation in the ether or a perceived correlation infected with subjective imprecision. In terms of the skilled addressee, he must be able to know or ascertain what is the correlation within claim 1 and what is the correlation without claim 1. Moreover, an invention where the correlation was not required to be statistically significant would, in my view, fail for lack of utility. The promise of the invention would be illusory and its fulfilment a fantasy.
Let me now make the following observations.
First, it is apparent that the claim as drafted includes any SNP covered by the claim that has any degree of association with any trait. In my view the claim lacks clarity because although a non-zero association might be determined by one study, it might be found not to have any association at all when tested using a different study. Moreover, the fact that the claim includes any degree of association also gives rise to lack of utility.
Second, even if the term “associated” requires a particular degree of association, which in my view would be to add an impermissible gloss, then the claim still lacks clarity. The person skilled in the art will not know the degree of statistical significance required to establish the requisite association. So in my opinion whether or not the claims require a particular degree of association, there is no workable standard in the claim for a person skilled in the relevant art to determine if a SNP that they are using is “associated” with a trait.
Now Branhaven confidently asserts that the measure of statistical significance or level of confidence is a matter that can readily be determined by a molecular geneticist. Now in one sense this is true. But in another sense it does not answer the lack of clarity question. Reasonable minds may differ as to the degree of association and statistical significance required. How is one to tell whether what one is doing falls inside or outside of claim 1? In my view claim 1 should be sufficiently clear on this point, which at the moment it is not.
In my view, statistical significance should be stipulated in a meaningful way and capable of objective assessment. Conformably with relevant aspects of the specification, a p value of equal to or less than 0.01 suggests itself; perhaps one could be more generous to Branhaven at a p value of equal to or less than 0.05, but I doubt it. I will leave that question open for further argument if Branhaven applies to amend claim 1. As to whether each of the 3 SNPs should separately satisfy the relevant p value or only the combination of the three, I will leave that for discussion on another occasion if necessary.
Finally and for completeness, I note that before the delegate, MLA had contended that there was a conflict in the specification regarding statistical thresholds for association. On the one hand, [0052] described an “associated SNP” as a SNP associated with a trait at a confidence level at 0.01 or greater. On the other hand, [0080] described SNPs associated with a trait as having statistical association of at least 80% to 99% confidence. Branhaven submitted that there was no conflict between the two paragraphs, because [0080] described a method of using a bovine SNP map to identify SNPs associated with traits. It was not seeking to define an “associated SNP”. The delegate was satisfied that [0052] related to the confidence level for identifying a SNP associated with a trait, whilst [0080] described a possible application of statistical methods to determine relationships between SNP nucleotide occurrences associated with traits. She construed the phrase “associated with a trait” to mean that the SNP was statistically determined to be significantly associated with a trait.
I have taken a slightly different approach. In my view that phrase should have been so expressed with some stipulation for the degree of statistical significance. But it has not been so expressed. Accordingly, the claim lacks clarity and does not properly define the invention absent some appropriate amendment to deal with my concern. I do not consider at all that it is appropriate to leave this to the skilled person. Some objective stipulation of statistical significance and the appropriate degree is necessary.
I will hear further from counsel on that question if Branhaven applies to amend.
(Original emphasis.)
331 Perhaps what I said in the second sentence of [297] could have been better expressed. But it was not at that point focused on and excluding the use of p-values. Indeed it is well apparent from later parts of my reasoning that the addition of p-values enhanced rather than detracted from clarity. And it seems clear that the main point that was being made was that the claims lacked clarity by reason of the absence of some stipulation for the degree of statistical significance. Finally, I reject an alternative argument made by MLA that the claims lack clarity if it covers “a situation where … after the trait is inferred without a step of determining a SNP trait association to the specified p-value, it is determined the SNPs are so associated”.
332 In summary I reject MLA’s other lack of clarity arguments.
(e) Conclusion
333 I reject MLA’s s 102 arguments. It is convenient now to turn to the question of discretion.
DISCRETION
334 I have accepted that I have power to entertain the amendment application and that the amendments are not disallowable by reason of s 102. Let me now turn to the question of the nature of the s 105(1A) power. Is the power discretionary or merely empowering? And if discretionary, what are the relevant factors to be considered?
335 Section 105(1A) provides, like s 105(1), that the court “may, on the application of the applicant for the patent [patentee], by order direct the amendment of the patent request or the complete specification in the manner specified in the order”. In my view, both provisions involve the application of a discretion.
336 Now Branhaven argued that the word “may” in s 105(1A) was merely empowering rather than discretionary. It contended that the context of s 105(1A), being an appeal from a decision of the Commissioner as a hearing de novo, was different to the context of an amendment under s 105(1) in proceedings for infringement or revocation. Further, Branhaven pointed out that under s 104, the Commissioner had no discretion not to allow an amendment. And by parity of reasoning it argued that as I was standing in the shoes of the Commissioner, equally I also had no discretion to allow the amendment provided, of course, that the amendments were not disallowable under s 102.
337 But at the time of s 105(1A)’s introduction, the explanatory memorandum stated “[e]xisting section 105 applies to amendment of patents. It is expected that in exercising their discretion under new subsection 105(1A), the courts will give account to different factors that are relevant to applications, in contrast to those applying to patents”. So, it was implicitly recognised that s 105(1A) conferred a discretionary power, albeit that different factors might inform the exercise of that discretion as compared with s 105(1).
338 Further, it is to be noted that in some respects the context in which the Court comes to hear an application to amend under either s 105(1) or s 105(1A) is not in substance different. Both involve an adversarial dispute between parties before the Court where the parties have greater rights in respect of the compulsion of the production of documents and the rules of engagement, whether procedural or evidentiary, concerning the forensic contest, than they would before the Commissioner. In essence, where a dispute about a patent application is unable to be determined at an administrative level, the matter is resolved by the exercise of federal judicial power. At that point, the dispute is the subject of a higher degree of scrutiny vis-à-vis attempts by the patent applicant to amend its patent application. The dispute is adjudicated in a forum designed to allow for the investigation, consideration and application of discretionary considerations including any relevant assessment of credit. Further, the exercise of judicial power may require consideration of a party’s conduct in terms of delay in seeking to invoke that power, the adequacy of disclosure to the Court, questions of election and procedural fairness, and effects on third parties by the exercise of judicial power. All of this suggests that “may” is discretionary rather than merely empowering under s 105(1A). But even if the power was not discretionary, it is difficult to see how a proper exercise of judicial power would prevent me from considering such matters in any event. No statute could by such conferral alone without further stipulation or express limitation necessarily exclude my consideration of such matters.
339 Further, the question of the amendment of a patent application in the context of an appeal to the Court from a decision of the Commissioner almost invariably arises in the context of a dispute about the validity of the patent application, which is in most respects similar to the proceedings to which s 105(1) applies.
340 Now I accept that if a dispute between a patentee and alleged infringer arises, the patentee is entitled to seek to apply pursuant to s 104 to amend its patent, and in that case the Commissioner will have no discretion not to amend for reasons that I will explain in a moment and assuming that s 102 is no bar. Similarly, if the alleged infringer asks the Commissioner to re-examine a granted patent and the patentee applies to amend the patent pursuant to s 104, no discretion arises. But should the dispute in either case proceed to Court before the amendment is made, for example because the alleged infringer asserts revocation, then the amendment application would need to be made to the Court and the Court has a discretion to refuse amendment. And if that is the case for patents, it is difficult to see any reason why the amendment of patent applications before a Court ought not to also involve an exercise of discretionary power, albeit that the relevant factors and balancing exercise might be different as compared with the amendment of granted patents.
341 Further and related to what I have just said, the position under s 104(2) does not provide any useful comparator. There is no commensurate discretion under s 104(2) because it requires the Commissioner to “consider and deal with the request [to amend] in accordance with the regulations”. The interaction between s 104(2) and Ch 10 of the Patents Regulations 1991(Cth) has the effect that the Commissioner’s power under s 104(2) is not discretionary (see New England Biolabs Inc v Commissioner of Patents (2001) 110 FCR 357 at [48] to [67] per Emmett J). But there is no such interaction which compels me to treat s 105(1A) in an analogous fashion to s 104(2). As I have indicated, I find it more compelling to treat s 105(1A) in a similar fashion to s 105(1) in the sense that both involve discretionary powers, albeit that the relevant factors and balancing exercise may overlap but are not completely co-extensive in their boundaries and content.
342 In summary, in my view the power under s 105(1A) is a discretionary power. Let me then turn to the relevant factors that ought inform the exercise of that discretion.
343 It is convenient to first refer to a few cases dealing with the exercise of discretion under s 105(1).
344 In Les Laboratoires Servier v Apotex Pty Ltd (2016) 247 FCR 61; [2016] FCAFC 27 the relevant factors were conveniently summarised at [242] to [245] by Bennett, Besanko and Beach JJ in the following terms:
While the power to amend should in appropriate circumstances be exercised in favour of the patentee, it bears mentioning that it will not always be possible to overcome a ground of revocation by an amendment. Accordingly, the ground of revocation sought to be overcome is also relevant to the way in which the discretion should be exercised. In the case of a failure to comply with the best method requirement, it would be necessary to take into account, in the exercise of the discretion, the reason for that obligation and the time at which it is meant to be fulfilled.
A number of principles have been established and discussed in detail in the authorities in the United Kingdom and in this Court with respect to amendment of granted patents, relevantly and in summary:
• The discretion exists for the benefit of the patentee.
• The onus to establish that amendment should be allowed is on the patentee.
• Generally, a permissible amendment (i.e. one which is permitted under the Act) will be allowed unless there are circumstances which would lead the court to refuse amendment.
• The patentee must make full disclosure of all relevant matters.
• The Court’s focus is on a patentee’s conduct, not the merit of an invention.
• Amendment should be sought promptly and where a patentee delays for an unreasonable period, the patentee has the onus of showing that it delayed on reasonable grounds, such as a belief, on reasonable grounds, that an amendment was not necessary.
• Unreasonable delay is a circumstance likely to lead to refusal of the amendment.
• In assessing delay, the time when the patentee was unaware and reasonably did not know of the need for amendment is not taken into account. The relevant delay is from when the patentee knows of the likely invalidity, or has its attention drawn to a defect in the patent, or is advised to strengthen the patent by amendment. That is, amendment will not be permitted in cases where a patentee knows or ought to know that amendment should be sought and fails to do so for a substantial period of time. Thus the reasonableness of the conduct of the patentee is a relevant consideration when assessing delay.
• Mere delay is not, of itself, sufficient to refuse to exercise the discretion to amend. The fact of delay is, however, relevant to whether the respondent or the general public have suffered detriment.
• If a patentee seeks to take unfair advantage of the unamended patent, knowing that it requires amendment, then refusal of the amendment is likely.
• The proportionality of the asserted culpability of the patentee as compared with the effect of loss of protection for the invention should be considered.
These principles have been applied to amendments generally and operate as factors relevant to the exercise of the discretion to grant or refuse amendments. There may be other factors that are relevant in a particular case.
We would add that the nature of the amendment and the ground of invalidity sought to be overcome by it are important matters to be taken into consideration when exercising the discretion.
345 Further, in an earlier decision between the same parties but concerning a different patent, the Full Court in Les Laboratoires Servier v Apotex Pty Ltd (2010) 273 ALR 630 at [92] per Kenny and Stone JJ observed that it was not necessary for the patentee to know that the unamended claims were invalid for delay to be a relevant factor in the exercise of the discretion. It was enough that the patentee knew of significant invalidity risks yet delayed in seeking amendment.
346 Further, a failure by the patentee to provide a full and frank disclosure of the reasons for seeking amendment is also a relevant factor in the exercise of the discretion. Further, where the patentee fails to voluntarily disclose an aspect of the reasons for seeking amendment until compelled to do so, that is also a relevant factor. Further, while obfuscation in the patentee’s explanation “may have been attributable in part to a desire to avoid waiver of client legal privilege, that possibility does not affect the fact that it was open to the primary judge to conclude that [the patentee] failed properly to disclose its reasons …” ([83]). Moreover, even if it is not possible for the patentee to disclose relevant facts without waiving privilege, that does not “transform [the patentee’s] obligation into something less than full and frank disclosure” (at [83]). Accordingly, I agree with MLA that the patentee cannot hide behind a claim of privilege as a reason for not providing full and frank disclosure.
347 In Novartis AG v Bausch & Lomb (Australia) Pty Ltd (2004) 62 IPR 71; [2004] FCA 835, after Merkel J allowed the deletion and consolidation of certain dependent claims, his Honour considered whether to allow an amendment to the original form of claim 1 to remove a typographical error and to add a limitation to make the claim conform with data that indicated the limits at which the invention would work dealing with ion permeability values. The invention that his Honour was concerned with was contact lenses involving a discovery concerning the correlation between ion permeability and “on eye” movement. There were two sets of values involved, one calculated by the ionoton measurement and the other calculated by the ionoflux measurement. The amendments were not disallowable under s 102. Accordingly his Honour considered the exercise of discretion. His Honour observed that the courts had identified a distinction between validating amendments that cure otherwise invalid claims (the case before me) and other amendments such as deletion amendments involving deleting invalid claims. At [66] his Honour said by reference to this distinction that:
… As explained above, the issue of whether the amendments are validating arguments can be relevant to the amendment application. The issue is not relevant because any higher standard of proof or higher standard of conduct is required to be established in respect of such amendments. Rather, the relevance relates to whether the more lenient approach that might be applied to deleting amendments, or amendments that merely seek to correct errors or mistakes or remove ambiguities might be applicable. The reason for the leniency was explained by Pearson LJ in Van der Lely at 74-5 where his Lordship observed that a court is prima facie disposed to the deletion of invalid claims because if such claims were to remain in the patent they would constitute a potential “nuisance to industry”. A similar observation can be made in respect of the mere correction of errors or mistakes which are not sought to cure any actual or potential invalidity.
(Original emphasis.)
348 On the question of whether full and frank disclosure had been made, the patentee asserted that certain figures which appeared in the claims were based on discussion, correspondence and thought by the inventors and scientific judgment. But there was no corroboration of those matters, which led to his Honour’s conclusion that the real reason behind the choice of the values in the claims (which were now sought to be amended) was “a policy on the part of [the patentee’s] attorneys to express the claims in the patent as broadly as possible, irrespective of whether the scientific evidence then available to the patentees supported those claims” (at [126]).
349 In the present case, MLA contends that no explanation or scientific justification has been offered by the patentee as to the original decision to claim SNPs within 500 kb either side of a limb (a) SNP, without any limitation as to linkage disequilibrium. But it seems well clear from my earlier discussion and indeed my principal reasons that LD was always treated as implicitly required, with the r2 metric and its degree now added to deal with the clarity concerns that I raised.
350 Merkel J also observed that while there is no difficulty with broad protection being claimed in a patent, the claims should “not travel beyond the matters disclosed in the specification and should not include embodiments of the invention that are known not to work or which cannot reasonably be expected to work” (at [128]). MLA says that the latter observation is particularly apposite to the present case. Perhaps it is, but I am not sure where it now goes given that I have disposed of MLA’s s 102 arguments.
351 His Honour concluded that the patentee had not made a full and accurate disclosure of all relevant matters “as it had not candidly disclosed the ‘whole story of how it was that amendment had come to be sought’” (at [130]).
352 His Honour then dealt with the question of delay. His Honour pointed out that amendment may be denied where the delay has caused detriment to a particular party or the general public. Moreover, where a lengthy delay has occurred, a reasonable explanation is required even if no public detriment is apparent (at [135]). In the case before him his Honour found that the patentee had been made aware of substantive errors in the claims six years before amendment was sought, but concluded that no proper explanation was given of the consideration given to the errors and why it was decided that no amendment was necessary.
353 In rejecting the amendments, his Honour observed at [161] to [162]:
The factors that militate against the granting of Novartis’ application to amend can be summarised as follows:
(i) the history of the patent’s drafting indicates that the ion permeability values contained in it were chosen not as a matter of scientific judgment but, rather, as part of a policy of claiming as broadly as possible, even if this meant overstating the patent’s claims without having reasonable grounds to do so;
(ii) when problems with the ion permeability values were revealed, no steps were taken to correct those values by the amendment of the patent until more than six years after the patentees were aware or ought to have been aware of the problems, and no reasonable or satisfactory explanation for that delay has been provided;
(iii) notwithstanding the problems concerning the ion permeability values, including the overstating of those values, there is no evidence that Novartis considered, or consulted its Australian legal advisers about, whether the patent should be amended until after the commencement of this proceeding and until after Bausch & Lomb had raised the issue of invalidity by reason of those ion permeability values;
(iv) the patentees have not made a full and candid disclosure to the court of all the circumstances relevant to the amendment application.
In summary, the present case is one in which the patentees knew or ought to have known that the substantive amendments they now seek to make should have been sought long ago, but have failed to provide a reasonable or satisfactory explanation for their delay, which can fairly be characterised as culpable delay. Further, the errors and other matters now sought to be corrected came about as part of a policy of overstating claims without having reasonable grounds for doing so.
354 Now this was a very different case to the case before me for reasons that will be apparent later when I discuss issues concerning knowledge of risk, delay and disclosure. Let me put the case law to one side and deal with some other points.
355 At the time of the proposed introduction of s 105(1A), the explanatory memorandum stated “[e]xisting section 105 applies to amendment of patents. It is expected that in exercising their discretion under new subsection 105(1A), the courts will give account to different factors that are relevant to applications, in contrast to those applying to patents”. Now MLA says that so much might be accepted for an unpublished and undisclosed patent application, but it says that as a matter of public policy there is no reason why the distinction should be otherwise made. It says that many of the matters identified by this Court as relevant to the application of the discretion under s 105 concern protection of the public against a patentee maintaining claims when it knows or ought to know that amendment of the claims should be sought and fails to do so for a substantial period of time. And this does not require the identification of any particular person who has read the published claims and acted to their detriment. This is because the claims stake out the monopoly granted to the patentee, the infringement of which grants the patentee extensive rights including pecuniary relief. Importantly, for the purposes of s 105(1A), s 57 provides that the right to pecuniary relief commences from the date of publication of the patent application:
After a complete specification relating to an application for a standard patent has become open to public inspection and until a patent is granted on the application, the applicant has the same rights as he or she would have had if a patent for the invention had been granted on the day when the specification became open to public inspection.
356 Now although such an action cannot be brought until grant, the public is on notice that relief can be sought by the patentee in respect of acts done from the date the specification was first published. The only caveat on that right is s 115, which provides that pecuniary relief may not be ordered in respect of any infringement of the patent made before amendment of the claims “unless the court is satisfied that the specification without the amendment was framed in good faith and with reasonable skill and knowledge”. Therefore, the public would be entitled to expect that the patent applicant would ensure that its claims were so drafted and properly mark out the boundaries of the proposed monopoly.
357 Now I must say that I am inclined to agree with these points.
358 Further, under s 105(1), the Court may reject an amendment where the patentee seeks to take advantage of patent claims that it knows or ought to know should be amended. But similar circumstances can arise with respect to a patent application where the patent applicant engages in negotiation and licensing of the technology the subject of the patent application (as in the present case). That is because, without the threat of that technology being the subject of granted patent claims, the licensee would have no need for a licence at all. And in such a context, it does not matter if the patent application is published or not, only whether the claims of the patent application are disclosed to the potential licensee and the right to exploit the claimed invention made the subject of negotiations. Moreover, the culpability of this behaviour is exacerbated where the patent applicant explicitly or implicitly threatens to sue for infringement on the grant of the patent claims, or makes some other like threat contingent on that grant, for example, threatening higher licence fees if a deal is not struck prior to grant.
359 Further, I accept that there is no good reason to distinguish between the amendment of patents and the amendment of patent applications in the need for full and frank disclosure. In my view, full and frank disclosure is required under s 105(1A) but in the context of the particular amendment application under consideration.
360 Contrastingly, Branhaven says that the factors identified as relevant to the exercise of the s 105(1) discretion are irrelevant to the exercise of the s 105(1A) discretion, including the question of delay. Indeed, it says in reliance on Mole Engineering that I should refrain from refusing the patent application where MLA’s objections can be cured by amendment.
361 Branhaven says that in the absence of guidance from the authorities, the factors relevant to the s 105(1A) discretion can be implied from the purposes of the relevant provisions in the Act, the explanatory memorandum and the Patents Regulations 1991 (Cth). It says that a principal purpose of the Act and the pre-grant procedures is to grant patent protection to patentable inventions where the requirements for patentability can be made out. Branhaven says that the principle in Mole Engineering is an expression of this purpose in allowing the opportunity for amendment to the claims of patent applications that disclose patentable inventions even if some or all of the claims as drafted are invalid.
362 Now I would reject these submissions to the extent that they are merely an alternative way of expressing its “no discretion” point that I have already rejected.
363 Now Branhaven points out that a Court cannot direct an amendment that is not allowable under s 102 (s 105(4)). And s 105(1A) does not specify any other factors a court should take into account when considering whether to direct the amendment of a patent application. Accordingly, as the Court is standing in the shoes of the Commissioner, Branhaven says that the first source of guidance should be the factors the Commissioner is directed to consider under the regulations. Pursuant to reg 10.5(1), if the amendments are proposed in anticipation of or in response to an examination report, the Commissioner must consider whether the proposed amendments would remove all lawful grounds of objection to the patent request and complete specification. Branhaven submits that response to or anticipation of an examination report disclosing lawful grounds of objection is a comparable circumstance to the circumstance where a Court has delivered reasons for judgment that identify lawful grounds of objection in an appeal from the decision of the Commissioner. Branhaven submits that apart from the statutory conditions in s 102, the factor of whether the amendments remove lawful grounds of objection should be the dominant consideration for the Court in exercising its discretion under s 105(1A) whether to direct the proposed amendments. If the proposed amendments do not remove the lawful grounds of objection identified in the Court’s reasons, then there is nothing achieved by directing the amendments. But if the proposed amendments are not disallowable pursuant to s 102, and also remove the identified grounds of objection, then the amendments should be directed and the patent application should proceed to grant.
364 Branhaven says that this approach is consistent with the reason for introducing s 105(1A), which is to reduce the complexity of the pre-grant process by empowering the Court to deal with amendment applications in an appeal from the Commissioner’s decision.
365 Now there is some force in these pellucid contentions put by Mr Christian Dimitriadis SC for Branhaven in the sense that I agree that the exercise of my discretion should take into account the matters just described. But where I part company from Branhaven’s position is that I disagree with Branhaven’s stark position that the following matters are irrelevant, being:
(a) knowledge of risk of potential difficulties;
(b) whether Branhaven has made full and frank disclosure; and
(c) unfair advantage and delay.
366 Let me flavour the discussion with a little nuance. I do not doubt that the three matters that I have just described are relevant to the exercise of my discretion under s 105(1A). But in the context of an application to amend a patent application as distinct from a granted patent, the context of my consideration of these matters, their weighting and the balancing exercise may have a different dimension as compared with considering such factors in exercising a discretion under s 105(1). And in the context of s 105(1A), as opposed to the context of s 105(1), the points made by Branhaven drawing an analogy between the Commissioner’s task and my task have significant relevance and weight in the discretionary balancing.
367 Finally on this aspect, Branhaven says that I should reject MLA’s argument that the Court cannot or should not afford an applicant an opportunity to amend as the Commissioner routinely does because the Court exercises judicial and not administrative power. The Court stands in the shoes of the Commissioner. Branhaven says that a consequence of MLA’s submission is that the powers of the Court are more limited than the powers of the Commissioner despite the express provision made in s 105(1A). It says that there is no support in the Act for such a proposition. But I think that MLA’s arguments and the relevant analysis is a little more subtle. Let me explain.
368 First, I am better placed than the Commissioner to adjudicate upon, and the Court’s processes are better able to facilitate an inquiry into, broader discretionary reasons. Second, the exercise of judicial power in the context of the assertion by Branhaven of an entitlement to amend (assuming s 102 is no bar) justifies my consideration of, inter-alia, any delay in invoking my judicial power, whether full and proper disclosure has been made to me to warrant a favourable exercise of judicial power, and whether third parties could be adversely affected by the exercise of my judicial power. In other words, if it be assumed that I am exercising judicial power (as it must be) in exercising a discretionary power (as I have so characterised it) under s 105(1A), then it is difficult to see how it could be said that such matters are irrelevant. The real question is rather one of the weighting of relevant considerations in the context of s 105(1A) as opposed to s 105(1).
369 Let me now turn to the discretionary grounds advanced that I consider it appropriate to take into account. It is convenient to deal with them in the sequence of:
(a) knowledge of risk of potential difficulties;
(b) whether Branhaven has made full and frank disclosure;
(c) unfair advantage and delay; and
(d) general matters.
(a) Knowledge of risk of potential difficulties
370 It is convenient at this point to set out some background concerning the history of the international applications. I will make the working assumption in favour of MLA that the knowledge of the relevant patent applicants and their assignees in relation to the international applications and the progress thereof is to be attributed to Branhaven and SelecTraits in addition to any direct knowledge that they may have.
371 The claims of the PCT application (PCT/US 2003/041766) as originally filed on 31 December 2003, insofar as they concerned claims to a method for inferring a trait of a bovine subject from a nucleic acid sample of the bovine subject, comprised identifying in the nucleic acid sample, at least one limb (a) SNP, wherein the limb (a) SNP is associated with the trait; but not all of the 2510 limb (a) SNPs were found to be associated with each of the five traits tested. Claim 1 referred only to a limb (a) SNP.
372 But claim 33 was to a method for identifying a bovine SNP associated with a trait which comprised identifying a “test SNP that is in linkage disequilibrium with” a limb (a) SNP. Claim 34 was to a method of claim 33, wherein the test region is less than or equal to about 500,000 nucleotides from a limb (a) SNP. Claim 34 was a claim to a test SNP that is in LD with a limb (a) SNP, which is associated with a trait, and which is a limb (b) SNP. It would appear that the patent applicants appreciated that for a test SNP to be useful it had to be in LD with a limb (a) SNP.
373 The PCT application made reference to the LD for cattle being about 500,000 nucleotides [0035]; see also [0126] and [0127].
374 The PCT application entered national phase and the claims were amended consistently in at least Australia, Canada and Europe so that claim 1 was no longer limited to the use of limb (a) SNPs. In response to the examiner’s first report that there was a lack of unity of the invention, the form adopted in the parent application on 17 November 2008 (a similar amendment from the PCT claims was made in the 253 Application on 29 April 2011), to reflect the position taken in Europe, was:
A method for identifying a trait of a bovine subject from a nucleic acid sample of the bovine subject, comprising identifying in the nucleic acid sample an occurrence of at least three single nucleotide polymorphisms (SNPs) wherein the SNPs are associated with the trait.
375 Claims 7 and 8 at that time were as follows:
7. The method of claim 1 or claim 2, wherein at least one SNP is located in a target region within 500,000 nucleotides of a trait associated SNP.
8. The method of claim 1 wherein the association is at a confidence interval of 0.01 or greater.
376 Claim 7 had the form of the limb (b) limitation except that the limb (b) SNP was not referable to a limb (a) SNP, but referable to any SNP that was found to be associated with the relevant trait. Claim 8 indicated an appreciation by the patent applicants that a claim to any association might be too broad and so required the identification of a particular level of association.
377 The concept of requiring linkage disequilibrium between a test SNP and an associated SNP was removed from the claims by these amendments by deleting claims 33 and 34. Now, according to MLA, the patent applicants are seeking to reintroduce such a requirement after having abandoned it in the 253 Application at least five years before the present amendment application. The amendment to remove linkage disequilibrium from the claims occurred in the 253 Application on 29 April 2011 (the parties incorrectly referred to this as 12 April 2012).
378 In Australia, on or around 1 September 2010, the patent applicants were aware that MLA objected to the grant of claim 10 of the parent application, which had the limb (b) requirement, on the basis that, inter-alia, the breadth of the claim rendered it invalid for lack of utility and clarity. MLA says that even accepting Dr Bentley’s contention that such complaints were really for an expert to opine on, the patent applicants were aware from on or around 18 May 2011 that Dr William Barendse held the view that “it is most unlikely that all SNPs within 500,000 nucleotides of any particular SNP would be useful in the same way as the defined SNP” (see his statutory declaration of 18 May 2011).
379 On 7 March 2012, in Europe, the examiner made observations concerning claim 7. Claim 7 added the requirement that “at least one SNP is located in a target region within 500,000 nucleotides of a trait associated SNP” to claim 1 which was to a method of inferring a trait in a bovine subject from a nucleic acid sample of that subject, comprising identifying from the sample three SNPs associated with the trait and occurring in genes. The European examiner said the following:
Furthermore, claim 7 implies that the SNPs to be tested are not necessarily associated to the desired trait, but can be in a 0.5 Mb region surrounding it. The reference to the independent claim is then contradictory and renders both claims unclear (Article 84 EPC). Moreover, this embodiment is based on the unsupported assumption that all SNPs within said distance of a marker SNP are also suitable markers of the desired trait, for the only reason of their “proximity” to the marker SNP. Considering the variability of linkage disequilibrium with factors like time, sample size, population, statistical tools, etc., and the presence of recombination hotspots distributed unevenly in the genome, this embodiment seems merely speculative (Article 83 EPC). The evidence presented in Example 3 could be considered convincing for the few SNPs tested, but generalization to all 2510 SNPs disclosed in the application appears unsupported in view of the above.
380 MLA says that the European examiner’s scientific concern was the same as was accepted to be true by Branhaven at trial. As recorded in my principal reasons “Branhaven says that … the skilled person would understand that not all SNPs within the region of +/- 500,000 nucleotides would be in LD with the specified SNP” (at [351]). I also accepted that without LD being specified in the claim the claim would lack utility.
381 On 17 September 2012, the patent applicants amended claim 7 in Europe to address the complaint that the SNP in claim 7 was not associated with the trait, but otherwise asserted that the examiner’s concerns were met by Example 3, which the examiner later rejected.
382 On 18 March 2013, in Canada, the examiner considered a claim in essentially the same form as being considered in Europe, where claim 10 in Canada had the limb (b) style requirement. The examiner made the same complaint as in Europe.
383 On 18 September 2013, the patent applicants by their submission in response to the Canadian examiner adopted a similar approach, with claim 1 reading:
A method for identifying a trait of a bovine subject from a nucleic acid sample of the bovine subject, comprising:
identifying in the nucleic acid sample an occurrence of at least three single nucleotide polymorphisms (SNPs)
wherein the at least three SNPs are associated with the trait, and wherein the at least three SNPs occur in more than one gene.
and claim 11 reading:
The method of any one of claims 1 to 8, wherein at least one SNP is located in a target region within 500,000 nucleotides of a trait associated SNP.
384 In Australia, on 30 April 2014 MLA filed its statement of grounds and particulars of objection to the grant of the 253 Application which had been accepted with the claims in their current form. As with the complaints made against claim 10 of the parent application, MLA raised grounds of utility, clarity and lack of definition.
385 On 9 May 2014, the European Patent Office (Examining Division) refused the European application at an oral hearing. The written decision was given on 26 May 2014. It was noted that in April 2014, the patent applicants had proposed auxiliary requests which sought to amend the claims to limit them to the limb (a) SNPs, then later to a single limb (a) SNP, which were the primary subject of the oral hearing and were also rejected.
386 On 20 May 2014, the Canadian examiner rejected claim 11 stating:
Claims 11 and 34 lack utility and do not comply with section 2 of the Patent Act. These claims imply that the SNPs to be tested can be in a 0.5 Mb region surrounding other SNPs. This embodiment is based on the unsupported assumption that all SNPs within said distance of a marker SNP are also suitable markers for the desired trait, only for the reason of their “proximity” to the marker SNP. Considering the variability of linkage disequilibrium with factors like time, sample size, population, statistical tools, etc., and the presence of recombination hotspots distributed unevenly in the genome, this embodiment seems merely speculative. The evidence presented in Example 3 could be considered convincing for the few SNPs tested, but generalization to all bovine SNPs disclosed lacks utility in view of the above.
Claims 11 and 34 are not fully supported by the description, and do not comply with section 84 of the Patent Rules. These claims imply that the SNPs to be tested can be in a 0.5 Mb region surrounding other SNPs. Applicant argues, in the letter dated September 18, 2013, that a skilled worker would have no difficulty in determining which SNPs are associated with the desired trait based on their proximity to the marker SNP. However, as stated in the office action dated March 18, 2013, this embodiment is based on the unsupported assumption that all SNPs within said distance of a marker SNP are also suitable markers for the desired trait, only for the reason of their “proximity” to the marker SNP. Considering the variability of linkage disequilibrium with factors like time, sample size, population, statistical tools, etc., and the presence of recombination hotspots distributed unevenly in the genome, this embodiment seems merely speculative. The evidence presented in Example 3 could be considered convincing for the few SNPs tested, but generalization to all bovine SNPs disclosed appears unsupported in view of the above.
387 On 28 August 2014, the patent applicants were advised by their Australian patent attorneys that while the Australian claims were not “so broad as to unduly risk invalidity” one “aspect of the claims that may be worth considering amending is the alternative requirement for a SNP within 500kb of position 300 of the specified sequences”. The advice was that this claim was “very broad, and possibly easier for another party to attack”. The patent attorneys proposed that an amendment be made to add further dependent claims omitting the option of a SNP being about 500kb from the identified sequences. The patent attorneys’ query in that regard was whether the patent applicants envisaged “the claims without the “500kb option” would also be commercially valuable in capturing the SNPs used by various companies”.
388 Now in my view it is appropriate to set out aspects of this email from Mr James Cherry:
1. I confirm the claims are of a “medium” scope as commented by Dan which is an effective compromise between being broad enough to catch an infringer, while not being so broad as to unduly risk invalidity. The likely strength of the claims if enforced in court is something we can get a clearer picture of while preparing evidence in the opposition as discussed.
2. One aspect of the claims that may be worth considering amending is the alternative requirement for a SNP within 500kb of position 300 of the specified sequences. This requirement is obviously very broad, and possibly easier for another party to attack. A simple way of improving your position would be to add further dependent claims identical to each of claims 1, 6, 7 and 8 but omitting the option of SNP being about 500kb from the identified sequences.
3. In terms of the proposed amendment above, do you envisage that claims without the “500kb option” would also be commercially valuable in capturing the SNPs used by various companies?
389 Now MLA says that this was all in the context of the same integer being the subject of complaint if not refusal in Australia, Canada and Europe and the subject of expert evidence from Dr Barendse in Australia. I will return to this email later.
390 Now MLA says that at the latest by 30 October 2014, Dr Bentley was aware that there was a dispute as to whether the claims of the 253 Application inherently required the limb (b) SNPs to be in LD with a limb (a) SNP, although I would note that she considered that question to be one for expert evidence. But MLA says that the fact that the claim did not expressly refer to LD was clear and the reliance on an inference from the language of the body of the specification ought to have been troubling to Dr Bentley. Regardless, MLA says that she appreciated that there was a divergence of scientific opinion as to whether the claims in Australia had an inherent requirement of LD, and accepted that there was a risk that it would be found that LD was not inherent in the claims. In the circumstances, MLA says that by 30 October 2014 at the latest, the patent applicants knew or ought reasonably to have known that there was a serious risk that the claims in Australia would be construed as not having an inherent requirement of LD.
391 On 20 November 2014, the patent applicants cancelled claim 11 (and claim 34) in Canada. But MLA says that Mr Smith failed to reveal this notwithstanding having directly referred to the 20 May 2014 Canadian examiner’s report and the patent applicants’ response to it, or to explain why claim 11 was cancelled. MLA says that it may be inferred that the patent applicants took the view that the Canadian examiner’s objections could not be overcome.
392 MLA says that neither in Canada nor Europe did the patent applicants take the position taken by their expert Professor Plastow in Australia that the limb (b) requirement inherently required the relevant LD. It says that such an assertion, if plausible, would have answered the scientific complaint made in Canada and Europe that proximity of SNPs were not predictive of utility because LD was so uncertain.
393 MLA says that there is no relevant distinction between claim 1 in Australia and claim 11 in Canada or claim 7 in Europe. The language used in all claims concerning the proximity of the unspecified SNP to the limb (a) SNP (in the Australian claims) or the associated SNP (in the Canadian claims) is the same. In Australia, claim 1 uses the phrase “wherein at least one SNP … is about 500,000 or less nucleotides from” a limb (a) SNP. In Canada, claim 11 used the phrase “wherein at least one SNP is located in a target region within 500,000 nucleotides of” the associated SNP. Similarly, the body of the specifications is not said to be different.
394 MLA says that it may be inferred that between 20 May and 20 November 2014, the patent applicants knew that the risk was significant given they did not assert, in response to the Canadian Examiner’s objection that claim 11 was inutile, that it inherently required LD on the same basis asserted by Professor Plastow in Australia.
395 Accordingly, MLA says that by November 2014 at the latest although arguably since 7 March 2012, the patent applicants knew or ought reasonably to have known that there was a significant risk that the 253 Application required amendment to include LD in some way to make the limb (b) SNP requirement useful and clear.
396 But I agree with Branhaven that MLA’s reliance on isolated events that took place during the Australian and various overseas prosecutions of analogous patent applications is selective and driven by hindsight.
397 In my view the most relevant jurisdiction is Australia. And MLA’s narrative disregards the significant fact that the examiner and the delegate relevantly upheld the claims in their present form. MLA’s chronology also disregards or mischaracterises the effect of the advice given by Dr Bentley in October 2013 and Mr Cherry in August 2014. This advice entailed in each case that the claims in their present form were not overly broad and could be maintained.
398 In evidence was an email chain dealing with various communications made by Dr Bentley on 15 and 16 October 2013 concerning potential concerns with the Australian claims. Various options were discussed by Dr Bentley in terms of degrees of likelihood of acceptance. But to me, this all seemed to be a plain vanilla analysis of what goes on behind the scenes concerning dealing with actual or potential issues. But no part of this email chain suggests to me that it was perceived at the time that the concerns that I have now raised were considered then to be likely knock out points. Indeed the main problem perceived at the time related to the unity question. Moreover, the discussion of the three options in the 15 October 2013 (11:01 pm) email and the outcome recorded in the 16 October 2013 (11.31 pm) email all more support Branhaven’s position I would have thought; see also Dr Bentley’s letter to the Commissioner of 17 October 2013.
399 Further, in relation to Mr Cherry’s email of 28 August 2014, parts of which have been reproduced earlier, although there was a discussion of limb (b) concerning breadth, the suggestion rather being made was not to remove limb (b) in its entirety but to add further dependent claims without the limb (b) aspect.
400 Further, the claims of the US patents demonstrate that those patents included claims that were considerably broader than those of the 253 Application as they stand, and as proposed to be amended. The US claims include claims that do not specify any particular SNPs. I agree with Branhaven that this substantially undermines MLA’s attempt to cherry pick from other jurisdictions (Europe and Canada) to support its proposition that Branhaven should have known that it needed to amend so as to narrow the Australian claims.
401 In relation to US patent no. 7,468,248, I would note the following:
(a) The form of claims as filed on 31 December 2003 included the following:
Claim 1:
A method for inferring a trait of a bovine subject from a nucleic acid sample of the bovine subject, comprising identifying in the nucleic acid sample, at least one nucleotide occurrence of at least one single nucleotide polymorphism (SNP) corresponding to position 300 of any one of SEQ ID NOS: 19473 to 21982, wherein the SNP is associated with the trait, thereby inferring the trait.
Claim 33:
A method for identifying a bovine single nucleotide polymorphism (SNP) associated with a trait, comprising identifying a test SNP that is in disequilibrium with a SNP position corresponding to position 300 of one of SEQ ID NOS: 19473 to 21982.
Claim 34:
The method of claim 33, wherein the test SNP is in a target region of a bovine genome, wherein the target region is within about 500,000 nucleotides from a SNP position corresponding to position 300 of one of SEQ ID NOS: 19473 to 21982.
(b) Amendments to those claims were filed on 2 April 2008 in the following form:
Claim 1:
A method for inferring a trait of a bovine subject from a nucleic acid sample of the bovine subject, comprising identifying in the nucleic acid sample, a nucleotide occurrence of a single nucleotide polymorphism (SNP) corresponding to position 300 of SEQ ID NO:21645, wherein the SNP is associated with the trait, thereby inferring the trait.
Claim 33:
A method for identifying a bovine single nucleotide polymorphism (SNP) associated with a trait, comprising identifying a test SNP that is in disequilibrium with a SNP position corresponding to position 300 of SEQ ID NO:21645.
Claim 34:
The method of claim 33, wherein the test SNP is in a target region of a bovine genome, wherein the target region is within about 500,000 nucleotides from a SNP position corresponding to position 300 of SEQ ID NO:21645.
402 In relation to US patent no. 7,709,206, I would note the following:
(a) The form of claims as filed on 13 February 2008 included the following:
Claims 1-100 cancelled
Claim 101:
A method for identifying a trait of an animal subject from a nucleic acid sample of the animal subject, comprising identifying in the nucleic acid sample an occurrence of at least three single nucleotide polymorphism (SNP) wherein the SNPs are associate with the trait.
Claim 112:
The method of claim 56 wherein at least one SNP is located in a target region within 500,000 nucleotides of a trait associated SNP.
Claim 113:
The method of claim 56 wherein the association is at a confidence interval of 0.01 or greater.
(b) Amendments to those claims were filed on 8 March 2010 in the following form:
Claim 101:
A method for identifying a trait of a bovine subject from a nucleic acid sample of the bovine subject, comprising
identifying in the nucleic acid sample an occurrence of at least three single nucleotide polymorphisms (SNPs),
wherein the at least three SNPs are associated with the trait, and
wherein the at least three SNPs occur in more than one gene.
Claim 112:
The method of claim 101, wherein at least one additional SNP is located in a target region within 500,000 nucleotides of a trait-associated SNP.
Claim 113:
The method of claim 101, wherein the association is at a confidence interval of 0.01 or greater.
403 In relation to US patent no. 8,450,064, I would note the following:
(a) The form of claims as filed on 8 March 2010 included the following:
Claim 1:
A method for inferring a trait of a bovine subject from a nucleic acid sample of the bovine subject, comprising identifying in the nucleic acid sample, at least one nucleotide occurrence of at least one single nucleotide polymorphism (SNP) corresponding to position 300 of any one of SEQ ID NOS: 19473 to 21982, wherein the SNP is associated with the trait, thereby inferring the trait.
Claim 33:
A method for identifying a bovine single nucleotide polymorphism (SNP) associated with a trait, comprising identifying a test SNP that is in disequilibrium with a SNP position corresponding to position 300 of one of SEQ ID NOS: 19473 to 21982.
Claim 34:
The method of claim 33, wherein the test SNP is in a target region of a bovine genome, wherein the target region is less than or equal to about 500,000 nucleotides from a SNP position corresponding to position 300 of one of SEQ ID NOS: 19473 to 21982.
(b) The form of claims as granted included the following:
Claim 101:
A method for matching a bovine trait-associated genotype with a bovine subject, comprising
amplifying at least 3 SNP regions in a nucleic acid sample from a bovine subject to form at least 3 amplification products, and identifying in each amplification product an occurrence of a single nucleotide polymorphism (SNP),
wherein the SNPs are in more than one gene or non-coding chromosomal region, and
wherein the trait-associated genotype is associated with a trait characteristic.
Claim 110:
The method of claim 101, wherein at least one additional SNP is located in a target region within 500,000 nucleotides of a trait-associated SNP.
404 In relation to US patent no. 9,206,478, I would note the following:
(a) Claim 101 was amended to the following on filing on 25 April 2013:
Claim 101:
A method of matching a bovine trait-associated genotype with a bovine subject comprising identifying in a nucleic acid sample from the bovine subject an occurrence of at least three single nucleotide polymorphisms (SNPs), wherein the at least three SNPs are associated with a trait, wherein the at least three SNPs occur in more than one gene, and sorting the bovine subject based on the associated trait.
(b) The US Attorney, Mr Giles made the following recommendations in relation to amending the pending application in his email to Mr Smith and Ms Skarohlid dated 20 February 2014:
“I therefore recommend some intermediate protection reciting the specific SNPs but without being restricted to any one specific SNP. The proposed claims are a repeat of the claims issued in U.S. Patent No. 8,450,064; however they further recite the specific SNPs listed in the original claims. Since the Examiner has already granted the broad claims in U.S. Patent No. 8,450,064, he should not restrict us to a specific SNP as occurred in the first patent. One of our patent agents, Janell Cleveland, knows this Examiner and can contact him to make sure that this is clear.
These amendments should overcome the statutory obviousness rejection. In addition, we have provided arguments to overcome the obviousness type double patent rejections. While these may not be successful, we recommend waiting to see if the Examiner maintains these rejections in view of the claim amendments before submitting terminal disclaimers”.
(c) Amendments to the claims were sought on 21 February 2014 in the following form:
Claim 101:
A method of matching a bovine trait-associated genotype with a bovine subject comprising identifying in a nucleic acid sample from the bovine subject an occurrence of at least three single nucleotide polymorphisms (SNPs), wherein at least one SNP corresponds to position 300 of any one of SEQ ID NOS: 19473 to 21982, wherein the at least three SNPs are associated with a trait, wherein the at least three SNPs occur in more than one gene, and sorting the bovine subject based on the associated trait.
Claim 111:
The method of claim 101, wherein at least one SNP is identified which is located in a target region within 500,000 nucleotides of a trait-associated SNP.
Claim 112:
The method of claim 101, wherein the genotype associated with a bovine trait has at least a 90% confidence interval.
(d) Amendments to the claims were sought on 8 April 2015 and the form of claims in the granted patent are the following:
Claim 101:
A method of matching a bovine trait-associated genotype with a bovine subject comprising assaying a nucleic acid sample from the bovine subject for an occurrence of at least three single nucleotide polymorphisms (SNPs), wherein at least one SNP corresponds to position 300 of any one of SEQ ID NOS: 19473 to 21982, wherein the at least three SNPs are associated with a trait, wherein the at least three SNPs occur in more than one gene.
Claim 111:
The method of claim 101, wherein at least one SNP is identified which is located in a target region within 500,000 nucleotides of a trait-associated SNP.
Claim 112:
The method of claim 101, wherein the genotype associated with a bovine trait has at least a 90% confidence interval.
405 Further, the fact that the US claims were referred to in an email as being “potentially at risk of attack for lack of written description and enablement” does not assist MLA. Those claims were examined, granted and still stand in that form. Further, it is obvious that a potential risk of attack does not equate to invalidity. Moreover, the requirements of written description and enablement differ considerably in Australia.
406 Further, it ought not to be forgotten that Branhaven has applied to amend in light of and in response to the findings made by me in my principal reasons, and to address the particular aspects of lack of clarity, definition and utility that I found to exist. Save for claim 13, which is not advanced by MLA as a basis for refusing the amendments, the first time these objections were upheld was by me in my reasons. The decision of the delegate from which the appeal was brought otherwise upheld the claims as they presently stand. And such claims had earlier been examined and approved by the Commissioner pursuant to the procedures under the Act.
407 Moreover, Branhaven’s parent application had been examined and approved by the Commissioner with the accepted claim 1 extending to the use of at least three SNPs without limitation to limb (a) and/or limb (b) SNPs. The limitation to SNPs within both limb (a) and limb (b) only arose in dependent claims 10 and 13. The limitation to limb (a) alone only arose in dependent claim 29.
408 Now MLA has relied on expert evidence filed by it in the opposition proceedings, which I might add that MLA did not choose to rely on in the appeal proceeding before me, to assert that upon receipt of this evidence, Branhaven should have amended the claims in the manner identified in my reasons; see the statutory declarations of Dr Barendse dated 18 May 2011 (filed in relation to the parent application) and 29 July 2014 (filed in relation to the opposition to the 253 Application). But the reality is that the delegate found that apart from claim 13, the claims were clear and useful.
409 MLA also relies on the amendments which were made on 29 April 2011 to assert that Branhaven should have amended the claims to include an express requirement for LD in the manner identified in my reasons. But the fact that the claims of the 253 Application were amended to delete a range of claims including the claim(s) in question does not support the proposition that Branhaven ought to have known that an express requirement for LD between the limb (a) and limb (b) SNPs was necessary for the claims to be valid. Moreover, the delegate found that the skilled person would understand that in the context of the disclosure of the specification, a SNP within 500kb of the reference SNP is equivalent to a SNP that is in LD with the reference SNP at position 300. That was also the position taken by Branhaven’s experts at the hearing of the appeal before me. In this context, I agree with Branhaven that it is unreasonable to suggest that the amendment of the claims at the earlier time indicates that Branhaven ought to have been aware then that an express requirement for LD was necessary.
410 Further, MLA’s reliance upon objections raised by examiners in other jurisdictions ignores the fact that the Australian examiners did not raise such objections in the prosecution of either the parent application or the 253 Application. Nor did the delegate find the claims of the 253 Application, other than claim 13, to be invalid.
411 In these circumstances, I agree with Branhaven that it was not incumbent upon it to undermine or “second guess” the result of the delegate’s decision, and the acceptance of the claims by the examiner, by abandoning those claims based on matters that arose during examination of patent applications in overseas jurisdictions, or in untested expert evidence filed by MLA in the opposition proceeding which was ultimately not accepted by the delegate.
412 Further, I tend to agree with Branhaven that an overall consideration of the evidence concerning the overseas jurisdictions more supports Branhaven’s position rather than MLA’s position. Claims of varying scope have been allowed in different jurisdictions, some broader than the claims of the 253 Application, for example claims not limited to any specified SNP or requiring only the use of “at least one” SNP, and some narrower.
413 Finally, the prosecution of patent applications is jurisdiction specific given the different laws and procedures that apply (Neurim Pharmaceuticals (1991) Ltd v Generic Partners Pty Ltd [2018] FCA 1082 at [46] to [48] and [52] per Nicholas J and Bayer Pharma Aktiengesellschaft v Generic Health Pty Ltd (2012) 99 IPR 59 at [210] and [213] per Yates J). It is problematic to say the least to glean much from the patent applicants’ overseas experience that is useful to assisting me to determine whether as an exercise of discretion I should refuse the amendment application. Now true it is that with the encouragement of MLA I have embarked upon a lengthy evidentiary odyssey over choppy foreign waters. But I should have stayed at home. Now let me be clear. Of course a patent applicants’ knowledge of risk that is relevant to an assessment of the amendment application may be informed by its overseas experiences, with the genesis being a PCT application, and its experiences thereafter being seen not as “a collection of isolated events, but as a continuum” (CSL Ltd v Novo Nordisk Pharmaceuticals Pty Ltd (No 2) (2010) 190 FCR 522 at [88] per Jessup J). But the relevant lens for the inquiry is knowledge of risk under Australian law, not foreign law. And judged through that lens, Branhaven is on the better side of the argument.
414 In summary, I do not consider that Branhaven had knowledge of a risk under Australian law that rose to the level of requiring it to act any earlier than it did in making the amendment application.
(b) Full and frank disclosure
415 MLA says that in order to discharge the onus upon it, Branhaven must place the “whole story” before the Court. MLA says that this requires Branhaven to have put forward evidence from witnesses who were able to speak with first-hand knowledge about all of the knowledge, understanding and conduct of Cargill and Branhaven relevant to the amendment application, as well as tendering all relevant documents. MLA says that such knowledge extends to matters which ought to have drawn Cargill’s and Branhaven’s attention to the possible need to amend to overcome a risk that the claims were too broad so as to be ill defined, inutile or lacking in clarity.
416 MLA also says that the requirement for full and frank disclosure means that the patentee may have to disclose documents that are privileged. But whether it does so is a matter for it. But it says that a failure to do so can entail that the amendment application should be refused.
417 MLA points out that Dr Bentley, Branhaven’s patent attorney gave evidence purporting to summarise the reasons for amendment. Further, Mr Smith, the intellectual property manager for Branhaven, gave evidence purporting to summarise relevant prosecution histories in Australia and overseas. But MLA says that no-one has been put forward to explain any of the following matters, which it says are relevant to the exercise of the discretion, being:
(a) how the claims came to be drafted in their presently invalid form;
(b) when the patent applicants first became aware that there was an issue (or complaint) that the form of the claims was too broad, or inutile, and what advice they sought or obtained and what decisions were made as a result;
(c) an explanation for the delay between the matters in (b) and the present amendment application; and
(d) the negotiations with MLA and others concerning the claims in their presently invalid form.
418 Further, it says that neither Cargill nor Branhaven have voluntarily disclosed any documents other than the publicly available documents annexed to the affidavits of Dr Bentley and Mr Smith. It says that the position of both Branhaven and Cargill has been that discovery was not necessary. Both Branhaven and Cargill contested MLA’s application for discovery and only provided documents relevant to discretion, apart from the publicly available documents referred to, when they were compelled to do so.
419 Further, MLA says that, significantly, both Cargill and Branhaven have chosen to claim privilege over their discovered documents except three publicly available documents in the case of Cargill. MLA says that the claim to privilege in circumstances where the question of the breadth of the claims and amendment of them was plainly in issue, combined with the failure of either Cargill or Branhaven to put forward a witness to tell the full story to me regarding amendment, fails to meet the obligation to make full and frank disclosure to support an amendment application.
420 Further, MLA says that Mr Smith’s attempt to characterise the (mere) production of documents as constituting disclosure was unconvincing, as was his preparedness to try and hide behind his attorneys on this issue. MLA says that the course of Mr Smith’s evidence is reflective of Branhaven’s dilatory attitude to disclosure, as shown by the “dogged” resistance to the production of documents until the “death knell” of trial, the “belated” waiver of privilege and even the silence about the assignment of Cargill’s interests until the last minute.
421 But regardless of the late production of documents, MLA says that it is now clear that Branhaven has not been full or frank in its disclosure of all relevant matters. It has referred to the following matters.
422 First, both Dr Bentley’s and Mr Smith’s affidavit evidence was directed to the issue of clarity when both had read my reasons and understood (or ought to have had pointed out to them) that my concerns about the claims extended beyond clarity into lack of definition and utility. Now while Dr Bentley’s focus on clarity is not explained by her, it is not suggested that she was personally aware of any contention by MLA that Branhaven had to give full and frank disclosure. But MLA says that Branhaven and its legal representatives knew that the need for full and frank disclosure was in issue. As to Mr Smith, MLA says that there is no excuse at all. He had read MLA’s particulars of objection to the amendment and knew that full and frank disclosure was in issue.
423 Second, MLA says generally that Mr Smith’s evidence about the position overseas was confined to whether concerns had been raised as to clarity that matched my concerns in that regard. In particular, MLA says that Mr Smith’s recitation of relevant matters in Europe and Canada was at best dissembling and, properly understood, misleading. It says that his evidence was intended to give me the impression that either there were no concerns raised that would have put Branhaven on notice of difficulties with the claims in Australia or that such problems had been overcome.
424 Third, in particular for Canada, MLA says that Mr Smith’s evidence gave the impression that concerns about a claim with a limb (b) style element only had an unrelated clarity issue raised about it that had been addressed by amendment and that only unrelated issues were raised in the Canadian examiner’s 20 May 2014 letter. But it says that the scientific concern regarding that claim was not in fact overcome and the claim Mr Smith identified as relevant (claim 10, which later became claim 11) was abandoned. MLA says that it cannot be that Mr Smith, or at least the persons who assisted him in drafting his evidence, was unaware of this given that he clearly read the relevant documents (the 20 May 2014 examiner’s report and the patent applicants’ response dated 20 November 2014).
425 Fourth, as for China, Mr Smith identified that “clarity issues in the terms identified in the reasons of Beach J … [had] not been raised in respect of the Chinese Parent Application or Chinese Divisional Application”. But MLA says that neither of those applications in fact have a claim that includes any kind of limb (b) style integer. MLA says that that is the disclosure that Mr Smith ought to have made.
426 Fifth, as for Europe, Mr Smith’s evidence was to the effect that, like in Canada, the objection to the claim including a limb (b) like integer had been overcome and unrelated issues had been raised when in fact the crucial and relevant objections according to MLA had been raised and maintained over years by the European examiner. MLA says that the European Patent Office in its reasons following an oral hearing had reported the patent applicants’ contentions regarding the claim in issue to be entirely unpersuasive and the patent applicants had instead proposed auxiliary claim sets which abandoned the claim in question.
427 Sixth, MLA says that the assignment of Cargill’s interests to SelecTraits does not assist Branhaven. As I said at the outset of my reasons, SelecTraits has taken an assignment of Cargill’s interest in the 253 Application. MLA says that the belated assignment from Cargill does not relieve SelecTraits or Branhaven from determining and then disclosing what Cargill knew and when about the likely invalidity of the claims. The assignment gave SelecTraits the right to call for all relevant documents from Cargill. Moreover, it is arguable that Cargill is under an implied obligation to provide such assistance as necessary to ensure that the assignment is not futile because of a failure on the amendment application to give full and frank disclosure.
428 Seventh, MLA says that there has been no evidence from Cargill as to what it knew and when about the vulnerability of the claims. Further, MLA says that due diligence is a standard feature of commercial transactions and should have been with respect to the assignment. But there has been no disclosure at all from SelecTraits. MLA says that in circumstances where SelecTraits seemingly has a number of connections with Branhaven, it may readily be inferred that SelecTraits must have known something about the 253 Application before it took the assignment, even if it did not get the information directly from Cargill. MLA says that given the commonality of interests, Branhaven’s knowledge should be imputed to SelecTraits. And given the assignment from Cargill to SelecTraits, Cargill’s knowledge should be imputed to SelecTraits.
429 In my view Branhaven has relevantly made the disclosure that I consider necessary for an exercise of discretion in its favour under s 105(1A). Moreover, many of MLA’s assertions in the context of the amendment application reacting to my concerns expressed in my principal reasons and where the amendments sought are not to a granted patent travel well beyond the requirements of any realistic full and frank disclosure into the territory of a counsel of perfection.
430 First, Branhaven called Dr Bentley to explain the nature and purpose of the amendments, and called both Dr Bentley and Mr Smith to provide details as to the prosecution of the 253 Application and the various applications in other jurisdictions. They provided detailed evidence and were made available for cross-examination.
431 Second, it is not such a grave sin that Mr Smith did not explain every aspect of the overseas prosecutions which MLA considers relevant. All of the relevant material was annexed to his affidavit or otherwise tendered. MLA was able to and did make what it could of that material. The relevant material was before me. Perhaps some aspects of his written evidence could have been more felicitously drafted (for example [5(o)] and [5(ff)], with also a better explanation that no limb (b) SNPs coverage was ultimately sought in Canada and China), but at the end of the day I consider that I had a reasonably complete picture of the salient facts necessary to inform my exercise of discretion in the present context.
432 Third, Branhaven gave discovery in the categories to which I found MLA was entitled, responded to a notice to produce, and waived privilege in some of the documents discovered and produced. Cargill, the former joint applicant, also waived privilege in documents it produced.
433 Fourth, there was some suggestion that Branhaven had failed to bring forward other relevant matters, such as a full explanation of the reasons why amendments were made elsewhere. But I agree with Branhaven that this goes well beyond anything that could be required in the present circumstances in the context before me. And Dr Bentley explained the reasons for the amendments proposed on the amendment application before me. They were, of course, to address the findings in my principal reasons.
434 Fifth, the advice to Branhaven that I have previously referred to does not assist MLA’s case. In particular, the email of 28 August 2014 from Mr Cherry did not suggest or recommend that the current claims should be narrowed or could not be pursued. To the contrary, Mr Cherry advised that the existing claims extending to limb (b) were “of a ‘medium’ scope” and “not … so broad as to unduly risk invalidity”. Further, he advised that a fall back option that “may be worth considering” involved “add[ing] further dependent claims” limited to limb (a) SNPs. But plainly this was on the basis that the existing broader claims extending to limb (b) SNPs could be maintained. Further, the advice from Dr Bentley in her emails of 15 and 16 October 2013 assists Branhaven. Dr Bentley provided three options concerning claims to which she considered Branhaven may be entitled. Branhaven went with the option Dr Bentley identified as the narrowest of those three, but the most likely to be accepted, reflecting the current claims. As Branhaven correctly submits, this did not involve covetous claiming, but rather acting on the advice of an experienced patent attorney in the relevant jurisdiction.
435 Sixth, as to the other perceived deficiencies raised by MLA including that there is no direct evidence from Cargill and SelecTraits, if there be any relevant non-disclosure it is of secondary importance at best in the present context. And I have taken it into account.
436 In summary, in the context of the s 105(1A) question, more than sufficient material was placed before me by Branhaven to justify a favourable exercise of discretion. Further, even if there was less than a full and frank disclosure, that is only a factor to consider in the exercise of discretion. In the present case it is not a bar to a favourable exercise of discretion, particularly in the context of s 105(1A).
(c) Unfair advantage and delay
437 MLA says that in 2014, during such time as Branhaven was in discussions with MLA and others over a potential commercial licence, Branhaven understood that there was a real risk with the claims, including the limb (b) SNPs, that they would be held not to be valid. In particular MLA says that Branhaven knew or ought reasonably to have known that the claims lacked utility, clarity and definition by reason of the claims merely referring to the proximity of one SNP to another within 500 kb when the inherent variability in LD over a genome gave such proximity no assured benefit to the claimed method.
438 MLA says that the patent applicants’ attorneys in Australia and Branhaven knew that claims limited to the limb (a) SNPs would not be as commercially valuable and that infringement of such claims would be easily avoided. Mr Cherry suspected that this was the case in his email advice to them of 28 August 2014. MLA says that Cargill also perceived that claims limited to the specific sequences could be easily worked around. Accordingly, MLA says that Branhaven and its attorneys understood that for there to be any commercial value at all in the claims of the 253 Application, it needed to maintain SNPs over and above the limb (a) SNPs. But MLA says that the options that had been presented to Branhaven in 2014, both by its Australian patent attorneys and its conduct overseas, in order to deal with the breadth of the limb (b) SNPs (in Australia) or more particularly the lack of utility arising from the limb (b) style integer (in Canada and Europe), was to limit the claims to the limb (a) SNPs (as suggested by Mr Cherry in Australia) or to in some other way abandon the limb (b) integer entirely (as occurred in Canada and Europe). It should be apparent from what I have said earlier that this submission is not based on a secure foundation. Branhaven was quite entitled to persist with its limb (b) SNP integer, subject of course to addressing the difficulties that I have identified in my principal reasons.
439 MLA says that it is in this context that Branhaven, admittedly in the business of making money from the licensing of its patents and patent applications, made the decision not to apply to amend the patent to address the problematic breadth of the limb (b) SNPs, but rather to maintain the broad claims so that it could pursue Branhaven’s strategy of monetising its assets. MLA says that Branhaven sought a licence in respect of the 253 Application in the form of the claims as they presently stand, but at the time it well knew or ought to have known of the significant risk with the claims.
440 MLA says that the fact that Branhaven’s Australian patent attorneys were happy to press ahead with the broad claims provides no real answer since, as Dr Bentley made clear, she had abrogated any responsibility as to matters of utility, clarity or definition to the patent applicants’ expert, Professor Plastow, and would have been content to proceed with the claims so long as they were arguably valid, even in the context of knowing that the broad claims were being asserted by Branhaven against MLA in licensing negotiations. MLA says that Dr Bentley accepted that as much as possible if there is an argument that a patent claim might be valid, she was happy to prosecute that claim, even where there might be a significant risk that it is not valid. This was after also accepting that it would have been in the patent applicants’ interests to maintain as broad a claim set as possible, while negotiating with MLA, regardless of their entitlement to the breadth of the claims. Indeed, MLA says that the Australian patent attorneys were equally aware that it was in the patent applicants’ commercial interests to maintain as broad a claim set as possible when negotiating with MLA and were attuned to prosecuting patents to maximise their client’s commercial strategies. I would note at this point that I do not consider MLA’s submissions to fairly reflect Dr Bentley’s evidence. What Dr Bentley said was that it primarily came down to what she believed the patent applicant was entitled to and that the claim scope she argued for, based on the information provided to her, was what she believed the applicant was entitled to. And as she said, “I will get the broadest claim scope that I believe they’re entitled”. And she accepted that such an approach “does typically place applicants in a stronger position for licensing”. I must say that I found that evidence to be unremarkable and such a practice entirely standard.
441 MLA says that the evidence establishes that although Branhaven understood and appreciated the significant risk that the claims to limb (b) SNPs may be invalid, and despite concerns over that limb of the claims being directly expressed by MLA, Branhaven chose not to amend and instead offered a commercial licence to MLA. I might say at this point that no licence was ever agreed.
442 MLA also says that “most strikingly” Mr Smith gave evidence that he did not take the advice presented to him by his Australian patent attorneys on 28 August 2014 to narrow the broad claims, because he was negotiating with MLA. MLA says that this was not because of any proper consideration of the significant risk of invalidity to the claims because of their extension to include limb (b) SNPs. MLA says that the evidence Mr Smith gave was generally to the effect that he did not give proper consideration to the validity concerns raised in various ways to limb (b) or any limb (b) style limitation to the claims. MLA contends that his and Branhaven’s primary driver was to maintain as broad a claim set as possible, regardless of concerns about validity, and in this case a significant risk as to lack of clarity, definition and utility. I would note at this point that I do not consider MLA’s submissions to fairly reflect Mr Smith’s evidence. As Mr Smith said under cross-examination, he believed that Mr Cherry advised “that we have a good case to proceed with them [i.e. the claims with the limb (b) SNPs]”. Further, even though Mr Smith agreed that the decision at the relevant time was not to apply to remove the limb (b) SNPs but rather to maintain the broad claims so that Branhaven could monetise its asset, so what? His state of belief was that they had a good case to proceed with them. Further, it ought not to be forgotten that in my principal reasons I did not reject the integer of limb (b) SNPs but rather stated that it was necessary to expressly refer to LD and to define a meaningful degree of LD. Moreover, Branhaven’s belief at all relevant times was that it considered that the LD requirement was implicit and would have been so understood by persons skilled in the art.
443 In summary, MLA submits that the evidence establishes that Branhaven sought to take unfair advantage of the unamended claims when it appreciated that there was a significant risk that the claims in their present form would be invalid.
444 I reject MLA’s ambitious submissions.
445 I agree with Branhaven that MLA has failed to adduce any evidence of unfair advantage. Ultimately, all that MLA sought to rely upon was that Branhaven had engaged in commercial negotiations with MLA as a result of MLA’s opposition to the parent application and the 253 Application. And as part of that negotiation, Branhaven sought a commercial resolution of the dispute involving the payment of licence fees.
446 Now one important context of these negotiations was that MLA demanded the complete deletion of limb (b) SNPs from the claims with the result that the claims would only claim the use of limb (a) SNPs. But I did not find that the deletion of limb (b) SNPs was required. In any event, MLA did not take a licence from Branhaven but sought to rely on its rights of pre-grant opposition and appeal. As a result, the 253 Application is still in the pre-grant phase. Neither MLA nor anyone else has been shown to have suffered any detriment or disadvantage.
447 Further, as to the cross-examination of Dr Bentley and Mr Smith on the issues concerning licensing activity, there is no evidence that Branhaven improperly or unfairly used the broader scope of the pending claims of the 253 Application, as compared with the claims as proposed to be amended, in order to extract licence fees or deter third parties from any activity. As Branhaven said, there is nothing improper in licensing activity per se. Further, there is nothing improper in seeking to secure the grant of the broadest claims to which a party may be entitled or to draft them, if permissible, in a manner that makes them difficult to design around; the Cargill email of 29 June 2011 in evidence was hardly a smoking gun. This is plain vanilla legitimate commercial activity for a patent applicant.
448 Moreover, as I have said, MLA did not take a licence. Further, the deal on which MLA was insisting, which involved limiting the claims to limb (a) SNPs, entailed Branhaven accepting claims which on the logic of MLA’s other contentions would have had no commercial value.
449 Finally, as to any failure to disclose Branhaven’s licensing activity, MLA abandoned its application for discovery of such material as part of a compromise proposal which enabled the tender of a bundle of inter partes correspondence. Further, MLA tendered various annexures to Ms McAlpine’s affidavit, although it did not read that affidavit. It appeared to me that MLA accepted that this material would be sufficient for it to be able to make its points on this issue. In context I do not accept that Branhaven had some additional obligation to provide more detailed evidence or disclosure on its licensing activity.
450 In summary, I do not consider that Branhaven has obtained any unfair advantage by delay. Indeed, I do not consider that Branhaven has unreasonably delayed in making the amendment application.
(d) General matters
451 Let me make some other points concerning the question of discretion.
452 First, the necessity for the amendments arises from my principal reasons, and the amendments are directly responsive to those reasons. Further and importantly, I did not say that the limb (b) SNPs integer had to be deleted. Rather I said that LD had to be made explicit, with the degree of LD stipulated. Many of MLA’s submissions on the amendment application seemed to somehow proceed on a different basis concerning the limb (b) SNPs integer including seeking to finesse much out of little in pointing out that the limb (b) SNP integer may not have been pressed in foreign jurisdictions. I have not said that it cannot be pressed in the 253 Application, indeed quite the opposite.
453 Second, even if there are some circumstances where the applicant for a patent should apply to amend the patent application prior to or at the hearing of the appeal before the Court, this case is not one of those circumstances.
454 Third, the so called “common law principles” referred to by MLA of election, Anshun estoppel, procedural fairness and more generally the proposition that a party is bound by its case do not advance MLA’s position. Branhaven did not elect to forgo any opportunity to amend. Further, there is no Anshun estoppel. More generally, it was not unreasonable for Branhaven not to seek the amendments prior to the commencement of the appeal or during the hearing thereof. The necessity for amendment only arose from my reasons. And the amendments are responsive to my reasons.
455 As I have said, prior to the delivery of my reasons, there was no finding that the three issues identified by me constituted lawful grounds of objection to the 253 Application.
456 No corresponding objections were raised by the examiner during examination of the 253 Application or by the Commissioner’s delegate during the hearing of the opposition.
457 Further, the 253 Application was the subject of an extensive opposition process that involved a significant amount of expert evidence followed by a hearing and then a detailed written decision from the Commissioner’s delegate. Aside from claim 13, that decision found that all of the claims as accepted were valid. Moreover, once MLA appealed the decision, it was for MLA to establish its opposition to the 253 Application in the de novo hearing before me, including in relation to claim 13.
458 Further, Branhaven at the hearing before me expressly reserved its right to apply to amend. Neither the election argument nor the Anshun estoppel argument have substance. Moreover, MLA has been accorded more than adequate procedural fairness. And as to the proposition that a party is bound by the conduct of its case, if it is accepted that I now have power to deal with the amendment application and, moreover, that I can permit amendments to address my concerns, the invocation of such a proposition in a context such as the present is superficial to say the least.
459 Fourth, acceptance of MLA’s approach would put patent applicants in a very difficult position. Where an opponent adopts a kitchen sink approach to use Branhaven’s description and raises numerous grounds of opposition, amending its notice of appeal several times in the course of doing so, a patent applicant would be forced to take the irreversible step of amending its claims prior to the evidence on which those grounds are based being tested at the hearing and prior to the grounds being considered by the Court on the merits. The alternative, on the logic of MLA’s argument, is to otherwise forgo any opportunity to amend. This could give the opponent a victory on grounds that might ultimately have had little merit, without them actually being determined by the Court. Moreover, it would discourage patent applicants from relying on legitimate and reasonably arguable defences to grounds of opposition, for fear of being found to be wrong and losing the patent application entirely. An amendment is, generally speaking, irreversible, in that once the claims have been narrowed, it is difficult to broaden them again (s 102(2)(b)).
460 In the present case, to the extent that MLA raised the relevant issues identified by me in my principal reasons in its grounds of opposition on appeal, these formed only a small part of the myriad of concerns raised by MLA in its appeal. I rejected the vast majority of its contentions. Further, the resolution of the contentions that were upheld by me depended on detailed expert evidence given at the hearing of the appeal. In my view Branhaven was entitled to await my findings before taking the irreversible step of amending the claims.
461 Fifth, the effect of MLA’s submissions is that a patent applicant who on the advice of its Australian attorneys seeks to prosecute a claim that has been upheld in examination and also upheld following a contested opposition before a delegate of the Commissioner, should be denied an opportunity to later amend on discretionary grounds because there was a perceived risk that the Court might ultimately reach a different view to the delegate, even if the position in support of the validity of the claims as they stood was reasonably arguable. To state the proposition is sufficient to justify its own rejection. But in any event, any commercially rational patent applicant would seek to maintain broader rather than narrower claims, if they could reasonably be supported, all other things being equal. There is nothing improper in such an approach or inappropriate about Branhaven waiting in the present case.
462 Sixth, I do not consider that Branhaven knew of significant likely invalidity risks of the type that I found. And in any event, even if it did this is only a discretionary factor to take into account. Moreover, the discretionary factors identified in the cases do not constitute some set of rigid rules to be inflexibly applied. And the discretion is a wide one.
463 Seventh, I am not here dealing with amendments to invalid claims in a granted patent and the context discussed by Emmett J in Les Laboratories Servier v Apotex Pty Ltd (2010) 273 ALR 630 at [57] in the following terms:
It is against the public interest for invalid claims in a patent to be maintained. Where a patentee seeks to maintain an invalid claim, in circumstances where the patentee knows or ought to know that the claim is invalid, the patent system is abused. Failure to amend on the basis of invalidity arising from lack of novelty, even after the patentee becomes aware of that invalidity, is relevant to the exercise of the discretion to permit amendment when subsequently applied for. On the other hand, if the patentee did not know nor ought to have known of the relevance of prior art and did not attempt to obtain some unfair advantage, a delay in seeking amendment will not bar the exercise of the discretion, so long as there is no unreasonable delay after the prior art comes to the attention of the patentee.
464 Eighth, the nature of the amendments sought in the present case are not irrelevant. For example, they are not designed to overcome prior art or to address a failure to disclose the best method where different considerations may apply. They are narrowing amendments to deal with clarity and definitional concerns raised by me. But I do accept that they are validating amendments save that the omission of claim 13 is a deletion amendment.
465 Ninth, and contrary to MLA’s protestations, I do not think that it can reasonably be said that Branhaven persisted with claims that it knew were likely to be unjustifiably wide. Further, it has not been shown that research and experiment has been deterred by the additional breadth. Further, it is important not to lose sight of the fact that I am dealing with an amendment to a patent application rather than a granted patent, although I accept the potential retrospective operation once any patent is granted (s 57) and that s 115(1) would not likely operate as a relevant restriction given that I am inclined to the view that the 253 Application (as unamended) was framed in good faith and with reasonable skill and knowledge; of course in the present context I am not deciding any s 115(1) question.
466 In summary and for the foregoing reasons, I propose to exercise my discretion in favour of allowing the amendments.
CONCLUSION
467 I will allow the amendments sought by Branhaven in the form set out in the schedule to these reasons. I will give the parties an opportunity to submit appropriate orders to deal with the amendments, final orders to deal with MLA’s appeal, and any additional submissions on costs.
468 In relation to costs I should make two points.
469 First, I have received some submissions already from the parties on the costs of MLA’s appeal. Subject to anything further that the parties wish to say on that point, I would propose that Branhaven pay 10% of MLA’s costs of that appeal. This is to reflect the fact that MLA had some modest success on its appeal and is a fair reflection of the issues on which it won. I reject Branhaven’s submission that it should have its costs or a substantial proportion thereof. MLA as the successful moving party, albeit in a modest way, is entitled to have some of its costs paid by Branhaven. I will make no order concerning Cargill’s costs one way or the other.
470 Second, on the costs of the amendment application I propose to make no order for costs in favour of any party. Branhaven was successful, but it obtained an indulgence. Further, on any view there had to be a hearing before me to address all relevant matters. Moreover, I was significantly assisted by MLA’s presentation which enabled me to proceed with the confidence that all relevant legal and forensic matters had been addressed. As to the Commissioner’s position as a non-party, I propose that Branhaven pay the Commissioner’s costs. But as I say, I will hear the parties further on any costs question, if necessary.
I certify that the preceding four hundred and seventy (470) numbered paragraphs are a true copy of the Reasons for Judgment herein of the Honourable Justice Beach. |
SCHEDULE – PROPOSED AMENDMENTS
THE CLAIMS DEFINING THE INVENTION ARE AS FOLLOWS:
1. A method for identifying a trait of a bovine subject from a nucleic acid sample of the bovine subject, comprising identifying in the nucleic acid sample an occurrence of at least three single nucleotide polymorphisms (SNPs) wherein each of the at least three SNPs are significantly associated with the trait, with the degree of statistical significance being p≤0.05, and wherein the at least three SNPs occur in more than one gene; and wherein
and wherein(a) at least one of the SNPs corresponds to position 300 of any one of SEQ ID NOS: 19473 to 21982, or
(b) the SNP is about 500,000 or less nucleotides from position 300 of any one of SEQ ID NOS: 19473 to 21982 and is in linkage disequilibrium with the SNP at position 300 with an r2 value of ≥0.7.
2. The method of claim 1, wherein the at least one SNP is selected from at least one of the SNPs and nucleotide occurrences indicated in Table 1A.
3. The method of claim 1 or 2, wherein occurrences of at least five SNPs are identified.
4. The method of claim 1 or 2, wherein occurrences of at least seven SNPs are identified.
5. The method of claim 1 or 2, wherein occurrences of at least ten SNPs are identified.
6. A method for sorting one or more bovine subjects, comprising:
a) identifying a trait for a first bovine subject from a nucleic acid sample of the first bovine subject, by a method comprising identifying an occurrence of at least three single nucleotide polymorphisms (SNPs) wherein each of the at least 3 SNPs are significantly associated with the trait, with the degree of statistical significance being p≤0.05, and the at least three SNPs occur in more than one gene, wherein at least one SNP corresponds to position 300 of at least one of SEQ ID NOS: 19473 to 21982, or the SNP is about 500,000 or less nucleotides from position 300 of any one of SEQ ID NOS: 19473 to 21982 and is in linkage disequilibrium with the SNP at position 300 with an r2 value of ≥ 0.7; and
b) sorting the first bovine subject based on the identified trait, and optionally repeating steps a) and b) for additional subjects.
7. A method for cloning a bovine subject with a desired trait, comprising:
a) identifying an occurrence of at least three single nucleotide polymorphisms (SNPs) wherein each of the at least 3 SNPs are significantly associated with the trait, with the degree of statistical significance being p≤0.05, and the at least three SNPs occur in more than one gene, wherein at least one SNP corresponds to position 300 of one of SEQ ID NOS: 19473 to 21982, or the SNP is about 500,000 or less nucleotides from position 300 of any one of SEQ ID NOS: 19473 to 21982 and is in linkage disequilibrium with the SNP at position 300 with an r2 value of ≥0.7;
b) isolating a progenitor cell from the bovine subject; and
c) generating a cloned bovine from the progenitor cell.
8. A method of inferring a trait in a bovine, comprising hybridizing a nucleic acid sample from a bovine subject to a system wherein the hybridizing comprises: a substrate; and specific binding pair members corresponding to a series of at least three SNPs, wherein each of the at least three SNPs are significantly associated with a trait in the bovine subject, with the degree of statistical significance being p≤0.05, and the at least three SNPs occur in more than one gene, wherein at least one of the SNPs corresponds to position 300 of any one of SEQ ID NOS: 19473 to 21982, or the SNP is about 500,000 or less nucleotides from position 300 of any one of SEQ ID NOS: 19473 to 21982 and is in linkage disequilibrium with the SNP at position 300 with an r2 value of ≥0.7,
where selective hybridization of the nucleic acid sample to the system indicates the presence of a SNP associated with a trait.
9. The method of any one of the preceding claims, wherein the trait is marbling, tenderness, quality grade, muscle content, fat thickness, feed efficiency, red meat yield, average daily weight gain, disease resistance, disease susceptibility, feed intake, protein content, bone content, maintenance energy requirement, mature size, amino acid profile, fatty acid profile, milk production, a milk quality susceptibility to the buller syndrome, stress susceptibility and response, temperament, digestive capacity, production of calpain, caplastatin and myostatin, pattern of fat deposition, ribeye area, fertility, ovulation rate, conception rate, fertility, or susceptibility to infection with and shedding of pathogens.
10. The method of any one of the preceding claims, wherein the trait is fat thickness, retail yield, tenderness, marbling, or average daily gain.
11. A bovine subject resulting from the method of claim 7.
12. A system for determining nucleotide occurrences of nucleotide polymorphisms (SNPs) in a bovine population comprising:
a hybridisation substrate that includes specific binding pair members corresponding to a series of at least 3 SNPs, wherein
the each of the SNPs in the series of SNPs are significantly associated with a trait from a bovine, with the degree of statistical significance being p≤0.05;
the SNPs occur in more than a single gene; and
at least one SNP corresponds to position 300 of any one of SEQ ID NOS: 19473 to 21982; or
the SNP is about 500,000 or less nucleotides from position 300 of any one of SEQ ID NO: 19473 to 21982 and is in linkage disequilibrium with the SNP at position 300 with an r2 value of ≥0.7.
13. An isolated polynucleotide identified according to the method of claim 8.
1413. An isolated polynucleotide when used in any one of methods 1 to 10 comprising:
a) a fragment of at least 20 contiguous nucleotides of a bovine genome, or
b) a complement of a);
wherein the isolated polynucleotide of a), or b), comprises a nucleotide occurrence of a single nucleotide polymorphism (SNP) significantly associated with a trait, with the degree of statistical significance being p≤0.05, wherein the SNP is about 500,000 or less nucleotides from position 300 of any one of SEQ ID NOS: 19473 to 21982 and is in linkage disequilibrium with the SNP at position 300 with an r2 value of ≥0.7, and wherein the isolated polynucleotide is less than or equal to about 500,000 nucleotides.
1514. The method of any one of claims 1 or 6 to 8, substantially as hereinbefore described.
15. A method for identifying a trait of a bovine subject from a nucleic acid sample of the bovine subject, comprising identifying in the nucleic acid sample an occurrence of at least three single nucleotide polymorphisms (SNPs) wherein each of the at least three SNPs are significantly associated with the trait, with the degree of statistical significance being p≤0.01, and wherein the at least three SNPs occur in more than one gene; and wherein
(a) at least one of the SNPs corresponds to position 300 of any one of SEQ ID NOS: 19473 to 21982, or
(b) the SNP is about 500.000 or less nucleotides from position 300 of any one of SEQ ID NOS: 19473 to 21982 and is in linkage disequilibrium with the SNP at position 300 with an r2 value of ≥0.7.
16. The method of claim 17, wherein the at least one SNP is selected from at least one of the SNPs and nucleotide occurrences indicated in Table 1A.
17. The method of claim 15 or 16, wherein occurrences of at least five SNPs are identified.
18. The method of claim 15 or 16, wherein occurrences of at least seven SNPs are identified.
19. The method of claim 15 or 16, wherein occurrences of at least ten SNPs are identified.
20. A method for sorting one or more bovine subjects, comprising:
a) identifying a trait for a first bovine subject from a nucleic acid sample of the first bovine subject, by a method comprising identifying an occurrence of at least three single nucleotide polymorphisms (SNPs) wherein each of the at least 3 SNPs are significantly associated with the trait, with the degree of statistical significance being p≤0.01, and the at least three SNPs occur in more than one gene, wherein at least one SNP corresponds to position 300 of at least one of SEQ ID NOS: 19473 to 21982, or the SNP is about 500,000 or less nucleotides from position 300 of any one of SEQ ID NOS: 19473 to 21982 and is in linkage disequilibrium with the SNP at position 300 with an r2 value of ≥0.7; and
b) sorting the first bovine subject based on the identified trait, and optionally repeating steps a) and b) for additional subjects.
21. A method for cloning a bovine subject with a desired trait, comprising:
a) identifying an occurrence of at least three single nucleotide polymorphisms (SNPs) wherein each of the at least 3 SNPs are significantly associated with the trait, with the degree of statistical significance being p≤0.01, and the at least three SNPs occur in more than one gene, wherein at least one SNP corresponds to position 300 of one of SEQ ID NOS: 19473 to 21982, or the SNP is about 500,000 or less nucleotides from position 300 of any one of SEQ ID NOS: 19473 to 21982 and is in linkage disequilibrium with the SNP at position 300 with an r2 value of ≥0.7;
b) isolating a progenitor cell from the bovine subject; and
c) generating a cloned bovine from the progenitor cell.
22. A method of inferring a trait in a bovine, comprising hybridizing a nucleic acid sample from a bovine subject to a system wherein the hybridizing comprises: a substrate; and specific binding pair members corresponding to a series of at least three SNPs, wherein each of the at least three SNPs are significantly associated with a trait in the bovine subject, with the degree of statistical significance being p≤0.01, and the at least three SNPs occur in more than one gene, wherein at least one of the SNPs corresponds to position 300 of any one of SEQ ID NOS: 19473 to 21982, or the SNP is about 500,000 or less nucleotides from position 300 of any one of SEQ ID NOS: 19473 to 21982 and is in linkage disequilibrium with the SNP at position 300 with an r2 value of ≥0.7,
where selective hybridization of the nucleic acid sample to the system indicates the presence of a SNP associated with a trait.
23. The method of any one of claims 15 to 22, wherein the trait is marbling, tenderness, quality grade, muscle content, fat thickness, feed efficiency, red meat yield, average daily weight gain, disease resistance, disease susceptibility, feed intake, protein content, bone content, maintenance energy requirement, mature size, amino acid profile, fatty acid profile, milk production, a milk quality susceptibility to the buller syndrome, stress susceptibility and response, temperament, digestive capacity, production of calpain, caplastatin and myostatin, pattern of fat deposition, ribeye area, fertility, ovulation rate, conception rate, fertility, or susceptibility to infection with and shedding of pathogens.
24. The method of any one of claims 15 to 23, wherein the trait is fat thickness, retail yield, tenderness, marbling, or average daily gain.
25. A bovine subject resulting from the method of claim 21.
26. A system for determining nucleotide occurrences of nucleotide polymorphisms (SNPs) in a bovine population comprising:
a hybridisation substrate that includes specific binding pair members corresponding to a series of at least 3 SNPs, wherein
each of the SNPs in the series of SNPs are significantly associated with a trait from a bovine, with the degree of statistical significance being p≤0.01;
the SNPs occur in more than a single gene; and
at least one SNP corresponds to position 300 of any one of SEO ID NOS: 19473 to 21982; or
the SNP is about 500,000 or less nucleotides from position 300 of any one of SEQ ID NO: 19473 to 21982 and is in linkage disequilibrium with the SNP at position 300 with an r2 value of ≥0.7.
27. An isolated polynucleotide when used in any one of methods 15 to 26 comprising:
a) a fragment of at least 20 contiguous nucleotides of a bovine genome, or
b) a complement of a);
wherein the isolated polynucleotide of a), or b), comprises a nucleotide occurrence of a single nucleotide polymorphism (SNP) significantly associated with a trait, with the degree of statistical significance being p≤0.01, wherein the SNP is about 500,000 or less nucleotides from position 300 of any one of SEQ ID NOS: 19473 to 21982 and is in linkage disequilibrium with the SNP at position 300 with an r2 value of ≥0.7, and wherein the isolated polynucleotide is less than or equal to about 500,000 nucleotides.
28. The method of any one of claims 15 or 20 to 22, substantially as hereinbefore described.
29. A method according to any one of claims 1 to 10, wherein the r2 value is ≥ 0.8.
30. A system according to claim 12, wherein the r2 value is ≥ 0.8.
31. An isolated polynucleotide according to claim 13, wherein the r2 value is ≥ 0.8.
32. A method according to any one of claims 15 to 24, wherein the r2 value is ≥ 0.8.
33. A system according to claim 26, wherein the r2 value is ≥ 0.8.
34. An isolated polynucleotide according to claim 27, wherein the r2 value is ≥ 0.8.




